Insulator protein Su(Hw) recruits SAGA and Brahma complexes and constitutes part of Origin Recognition Complex-binding sites in the Drosophila genome
- PMID: 23609538
- PMCID: PMC3675495
- DOI: 10.1093/nar/gkt297
Insulator protein Su(Hw) recruits SAGA and Brahma complexes and constitutes part of Origin Recognition Complex-binding sites in the Drosophila genome
Abstract
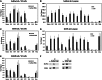
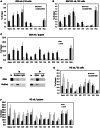
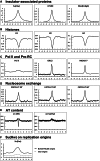
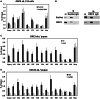
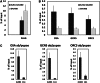
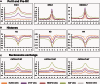
Despite increasing data on the properties of replication origins, molecular mechanisms underlying origin recognition complex (ORC) positioning in the genome are still poorly understood. The Su(Hw) protein accounts for the activity of best-studied Drosophila insulators. Here, we show that Su(Hw) recruits the histone acetyltransferase complex SAGA and chromatin remodeler Brahma to Su(Hw)-dependent insulators, which gives rise to regions with low nucleosome density and creates conditions for ORC binding. Depletion in Su(Hw) leads to a dramatic drop in the levels of SAGA, Brahma and ORC subunits and a significant increase in nucleosome density on Su(Hw)-dependent insulators, whereas artificial Su(Hw) recruitment itself is sufficient for subsequent SAGA, Brahma and ORC binding. In contrast to the majority of replication origins that associate with promoters of active genes, Su(Hw)-binding sites constitute a small proportion (6%) of ORC-binding sites that are localized preferentially in transcriptionally inactive chromatin regions termed BLACK and BLUE chromatin. We suggest that the key determinants of ORC positioning in the genome are DNA-binding proteins that constitute different DNA regulatory elements, including insulators, promoters and enhancers. Su(Hw) is the first example of such a protein.
Figures
References
-
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. - PubMed
-
- Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell. 2004;16:737–748. - PubMed
-
- Kurshakova M, Maksimenko O, Golovnin A, Pulina M, Georgieva S, Georgiev P, Krasnov A. Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol. Cell. 2007;27:332–338. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

