Exploring mRNA 3'-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening
- PMID: 23609544
- PMCID: PMC3675481
- DOI: 10.1093/nar/gkt265
Exploring mRNA 3'-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening
Abstract
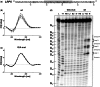
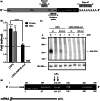
Guanine-rich RNA sequences can fold into non-canonical, four stranded helical structures called G-quadruplexes that have been shown to be widely distributed within the mammalian transcriptome, as well as being key regulatory elements in various biological mechanisms. That said, their role within the 3'-untranslated region (UTR) of mRNA remains to be elucidated and appreciated. A bioinformatic analysis of the 3'-UTRs of mRNAs revealed enrichment in G-quadruplexes. To shed light on the role(s) of these structures, those found in the LRP5 and FXR1 genes were characterized both in vitro and in cellulo. The 3'-UTR G-quadruplexes were found to increase the efficiencies of alternative polyadenylation sites, leading to the expression of shorter transcripts and to possess the ability to interfere with the miRNA regulatory network of a specific mRNA. Clearly, G-quadruplexes located in the 3'-UTRs of mRNAs are cis-regulatory elements that have a significant impact on gene expression.
Figures


References
-
- Huppert JL. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008;37:1375–1384. - PubMed
-
- Neidle S, Balasubramanian S. Quadruplex Nucleic Acids. Cambridge: RSC Publishing; 2006. p. 301.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

