Ldb1-nucleated transcription complexes function as primary mediators of global erythroid gene activation
- PMID: 23610375
- PMCID: PMC3668490
- DOI: 10.1182/blood-2013-01-479451
Ldb1-nucleated transcription complexes function as primary mediators of global erythroid gene activation
Abstract
Erythropoiesis is dependent on the lineage-specific transcription factors Gata1, Tal1, and Klf1. Several erythroid genes have been shown to require all 3 factors for their expression, suggesting that they function synergistically; however, there is little direct evidence for widespread cooperation. Gata1 and Tal1 can assemble within higher-order protein complexes (Ldb1 complexes) that include the adapter molecules Lmo2 and Ldb1. Ldb1 proteins are capable of coassociation, and long-range Ldb1-mediated oligomerization of enhancer- and promoter-bound Ldb1 complexes has been shown to be required for β-globin gene expression. In this study, we generated a genomewide map of Ldb1 complex binding sites that revealed widespread binding at erythroid genes and at known erythroid enhancer elements. Ldb1 complex binding sites frequently colocalized with Klf1 binding sites and with consensus binding motifs for other erythroid transcription factors. Transcriptomic analysis demonstrated a strong correlation between Ldb1 complex binding and Ldb1 dependency for gene expression and identified a large cohort of genes coregulated by Ldb1 complexes and Klf1. Together, these results provide a foundation for defining the mechanism and scope of Ldb1 complex activity during erythropoiesis.
Figures
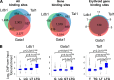
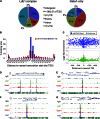



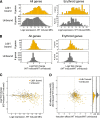
References
-
- Schlaeger TM, Mikkola HK, Gekas C, Helgadottir HB, Orkin SH. Tie2Cre-mediated gene ablation defines the stem-cell leukemia gene (SCL/tal1)-dependent window during hematopoietic stem-cell development. Blood. 2005;105(10):3871–3874. - PubMed
-
- Pevny L, Simon MC, Robertson E, et al. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349(6306):257–260. - PubMed
-
- Welch JJ, Watts JA, Vakoc CR, et al. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104(10):3136–3147. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

