Rhomboid domain-containing protein 3 is a negative regulator of TLR3-triggered natural killer cell activation
- PMID: 23610400
- PMCID: PMC3651449
- DOI: 10.1073/pnas.1220466110
Rhomboid domain-containing protein 3 is a negative regulator of TLR3-triggered natural killer cell activation
Abstract
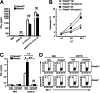
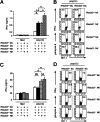
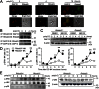
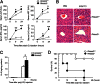
Rhomboid domain-containing protein 3 (Rhbdd3), which belongs to a family of proteins with rhomboid domain, is widely expressed in immune cells; however, the roles of the Rhbdd members, including Rhbdd3, in immunity remain unknown. Natural killer (NK) cells are critical for host immune defense and also can mediate inflammatory diseases such as hepatitis. Although much is known about how NK cells are activated, the detailed mechanisms for negative regulation of NK cell activation remain to be fully understood. Using Rhbdd3-deficient mice, we reveal that Rhbdd3, selectively up-regulated in NK cells upon Toll-like receptor 3 (TLR3) stimulation, negatively regulates TLR3-mediated NK cell activation in a feedback manner. Rhbdd3 inhibits TLR3-triggered IFN-γ and granzyme B expression of NK cells in cell-cell contact dependence of accessory cells such as dendritic cells and Kupffer cells. Rhbdd3 interacts with DNAX activation protein of 12 kDa and promotes its degradation, inhibiting MAPK activation in TLR3-triggered NK cells. Furthermore, Rhbdd3 plays a critical role in attenuating TLR3-triggered acute inflammation by controlling NK cell activation and accumulation in liver and disrupting NK cell-Kupffer cell interaction. Therefore, Rhbdd3 is a feedback inhibitor of TLR3-triggered NK cell activation. Our study outlines a mechanism for the negative regulation of NK cell activation and also provides clues for the function of the rhomboid proteins in immunity.
Keywords: immune regulation; innate immunity; poly(I:C).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Tyrosine kinase Btk is required for NK cell activation.J Biol Chem. 2012 Jul 6;287(28):23769-78. doi: 10.1074/jbc.M112.372425. Epub 2012 May 15. J Biol Chem. 2012. PMID: 22589540 Free PMC article.
-
TLR3 and Rig-like receptor on myeloid dendritic cells and Rig-like receptor on human NK cells are both mandatory for production of IFN-gamma in response to double-stranded RNA.J Immunol. 2010 Aug 15;185(4):2080-8. doi: 10.4049/jimmunol.1000532. Epub 2010 Jul 16. J Immunol. 2010. PMID: 20639488 Free PMC article.
-
The beta2 integrin CD11b attenuates polyinosinic:polycytidylic acid-induced hepatitis by negatively regulating natural killer cell functions.Hepatology. 2009 Nov;50(5):1606-16. doi: 10.1002/hep.23168. Hepatology. 2009. PMID: 19821527
-
TLR3: interferon induction by double-stranded RNA including poly(I:C).Adv Drug Deliv Rev. 2008 Apr 29;60(7):805-12. doi: 10.1016/j.addr.2007.11.005. Epub 2008 Jan 2. Adv Drug Deliv Rev. 2008. PMID: 18262679 Review.
-
FAM26F: An Enigmatic Protein Having a Complex Role in the Immune System.Int Rev Immunol. 2023;42(4):247-257. doi: 10.1080/08830185.2016.1206098. Epub 2016 Sep 19. Int Rev Immunol. 2023. PMID: 27645024 Review.
Cited by
-
Comparative Analysis of MicroRNA and mRNA Profiles of Sperm with Different Freeze Tolerance Capacities in Boar (Susscrofa) and Giant Panda (Ailuropodamelanoleuca).Biomolecules. 2019 Sep 1;9(9):432. doi: 10.3390/biom9090432. Biomolecules. 2019. PMID: 31480517 Free PMC article.
-
Genome-wide association and transcriptome studies identify target genes and risk loci for breast cancer.Nat Commun. 2019 Apr 15;10(1):1741. doi: 10.1038/s41467-018-08053-5. Nat Commun. 2019. PMID: 30988301 Free PMC article.
-
Cellular and molecular regulation of innate inflammatory responses.Cell Mol Immunol. 2016 Nov;13(6):711-721. doi: 10.1038/cmi.2016.58. Epub 2016 Oct 31. Cell Mol Immunol. 2016. PMID: 27818489 Free PMC article. Review.
-
Substrate binding and specificity of rhomboid intramembrane protease revealed by substrate-peptide complex structures.EMBO J. 2014 Oct 16;33(20):2408-21. doi: 10.15252/embj.201489367. Epub 2014 Sep 12. EMBO J. 2014. PMID: 25216680 Free PMC article.
-
Rhomboids, signalling and cell biology.Biochem Soc Trans. 2016 Jun 15;44(3):945-50. doi: 10.1042/BST20160035. Biochem Soc Trans. 2016. PMID: 27284064 Free PMC article. Review.
References
-
- Yokoyama WM. Mistaken notions about natural killer cells. Nat Immunol. 2008;9(5):481–485. - PubMed
-
- Baxter AG, Smyth MJ. The role of NK cells in autoimmune disease. Autoimmunity. 2002;35(1):1–14. - PubMed
-
- Mondelli MU, Varchetta S, Oliviero B. Natural killer cells in viral hepatitis: Facts and controversies. Eur J Clin Invest. 2010;40(9):851–863. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

