Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells
- PMID: 23610439
- PMCID: PMC3651503
- DOI: 10.1073/pnas.1213431110
Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells
Abstract
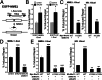
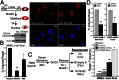
Microhomology-mediated end joining (MMEJ) is a major pathway for Ku-independent alternative nonhomologous end joining, which contributes to chromosomal translocations and telomere fusions, but the underlying mechanism of MMEJ in mammalian cells is not well understood. In this study, we demonstrated that, distinct from Ku-dependent classical nonhomologous end joining, MMEJ--even with very limited end resection--requires cyclin-dependent kinase activities and increases significantly when cells enter S phase. We also showed that MMEJ shares the initial end resection step with homologous recombination (HR) by requiring meiotic recombination 11 homolog A (Mre11) nuclease activity, which is needed for subsequent recruitment of Bloom syndrome protein (BLM) and exonuclease 1 (Exo1) to DNA double-strand breaks (DSBs) to promote extended end resection and HR. MMEJ does not require S139-phosphorylated histone H2AX (γ-H2AX), suggesting that initial end resection likely occurs at DSB ends. Using a MMEJ and HR competition repair substrate, we demonstrated that MMEJ with short end resection is used in mammalian cells at the level of 10-20% of HR when both HR and nonhomologous end joining are available. Furthermore, MMEJ is used to repair DSBs generated at collapsed replication forks. These studies suggest that MMEJ not only is a backup repair pathway in mammalian cells, but also has important physiological roles in repairing DSBs to maintain cell viability, especially under genomic stress.
Keywords: BLM/Exo1; CtIP; DNA damage; DNA repair pathway; genome stability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


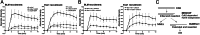

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

