Control of cell proliferation, endoreduplication, cell size, and cell death by the retinoblastoma-related pathway in maize endosperm
- PMID: 23610440
- PMCID: PMC3651506
- DOI: 10.1073/pnas.1304903110
Control of cell proliferation, endoreduplication, cell size, and cell death by the retinoblastoma-related pathway in maize endosperm
Abstract
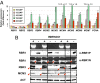
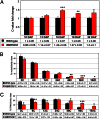
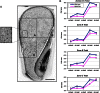
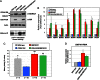
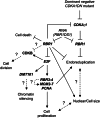
The endosperm of cereal grains is one of the most valuable products of modern agriculture. Cereal endosperm development comprises different phases characterized by mitotic cell proliferation, endoreduplication, the accumulation of storage compounds, and programmed cell death. Although manipulation of these processes could maximize grain yield, how they are regulated and integrated is poorly understood. We show that the Retinoblastoma-related (RBR) pathway controls key aspects of endosperm development in maize. Down-regulation of RBR1 by RNAi resulted in up-regulation of RBR3-type genes, as well as the MINICHROMOSOME MAINTENANCE 2-7 gene family and PROLIFERATING CELL NUCLEAR ANTIGEN, which encode essential DNA replication factors. Both the mitotic and endoreduplication cell cycles were stimulated. Developing transgenic endosperm contained 42-58% more cells and ∼70% more DNA than wild type, whereas there was a reduction in cell and nuclear sizes. In addition, cell death was enhanced. The DNA content of mature endosperm increased 43% upon RBR1 down-regulation, whereas storage protein content and kernel weight were essentially not affected. Down-regulation of both RBR1 and CYCLIN DEPENDENT KINASE A (CDKA);1 indicated that CDKA;1 is epistatic to RBR1 and controls endoreduplication through an RBR1-dependent pathway. However, the repressive activity of RBR1 on downstream targets was independent from CDKA;1, suggesting diversification of RBR1 activities. Furthermore, RBR1 negatively regulated CDK activity, suggesting the presence of a feedback loop. These results indicate that the RBR1 pathway plays a major role in regulation of different processes during maize endosperm development and suggest the presence of tissue/organ-level regulation of endosperm/seed homeostasis.
Keywords: endocycle; seed development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Sabelli PA, Larkins BA. The contribution of cell cycle regulation to endosperm development. Sex Plant Reprod. 2009;22(4):207–219. - PubMed
-
- Larkins BA, et al. Investigating the hows and whys of DNA endoreduplication. J Exp Bot. 2001;52(355):183–192. - PubMed
-
- Sabelli PA. Replicate and die for your own good: Endoreduplication and cell death in the cereal endosperm. J Cereal Sci. 2012;56(1):9–20.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

