Design and Development of Artificial Zinc Finger Transcription Factors and Zinc Finger Nucleases to the hTERT Locus
- PMID: 23612114
- PMCID: PMC3650244
- DOI: 10.1038/mtna.2013.12
Design and Development of Artificial Zinc Finger Transcription Factors and Zinc Finger Nucleases to the hTERT Locus
Abstract
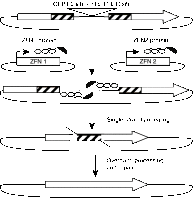
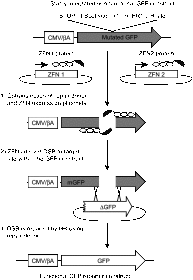
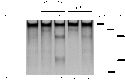
The ability to direct human telomerase reverse transcriptase (hTERT) expression through either genetic control or tunable regulatory factors would advance not only our understanding of the transcriptional regulation of this gene, but also potentially produce new strategies for addressing telomerase-associated disease. In this work, we describe the engineering of artificial zinc finger transcription factors (ZFTFs) and ZF nucleases (ZFNs) to target sequences within the hTERT promoter and exon-1. We were able to identify several active ZFTFs that demonstrate a broadly tunable response when screened by a cell-based transcriptional reporter assay. Using the same DNA-binding domains, we generated ZFNs that were screened in combinatorial pairs in cell-based extrachromosomal single-strand annealing (SSA) assays and in gene-targeting assays using stably integrated constructs. Selected ZFN pairs were tested for the ability to induce sequence changes in a Cel1 assay and we observed frequencies of genomic modification up to 18.7% at the endogenous hTERT locus. These screening strategies have pinpointed several ZFN pairs that may be useful in gene editing of the hTERT locus. Our work provides a foundation for using engineered ZF proteins (ZFPs) for modulation of the hTERT locus.Molecular Therapy - Nucleic Acids (2013) 2, e87; doi:10.1038/mtna.2013.12; published online 23 April 2013.
Figures


Similar articles
-
Genome editing with CompoZr custom zinc finger nucleases (ZFNs).J Vis Exp. 2012 Jun 14;(64):e3304. doi: 10.3791/3304. J Vis Exp. 2012. PMID: 22732945 Free PMC article.
-
Simultaneous screening and validation of effective zinc finger nucleases in yeast.PLoS One. 2013 May 31;8(5):e64687. doi: 10.1371/journal.pone.0064687. Print 2013. PLoS One. 2013. PMID: 23741369 Free PMC article.
-
Expanding the Repertoire of Target Sites for Zinc Finger Nuclease-mediated Genome Modification.Mol Ther Nucleic Acids. 2013 Apr 30;2(4):e88. doi: 10.1038/mtna.2013.13. Mol Ther Nucleic Acids. 2013. PMID: 23632390 Free PMC article.
-
Origins of Programmable Nucleases for Genome Engineering.J Mol Biol. 2016 Feb 27;428(5 Pt B):963-89. doi: 10.1016/j.jmb.2015.10.014. Epub 2015 Oct 23. J Mol Biol. 2016. PMID: 26506267 Free PMC article. Review.
-
Use of Zinc-Finger Nucleases for Crop Improvement.Prog Mol Biol Transl Sci. 2017;149:47-63. doi: 10.1016/bs.pmbts.2017.03.006. Epub 2017 Apr 29. Prog Mol Biol Transl Sci. 2017. PMID: 28712500 Review.
Cited by
-
Orthogonal Cas9-Cas9 chimeras provide a versatile platform for genome editing.Nat Commun. 2018 Nov 19;9(1):4856. doi: 10.1038/s41467-018-07310-x. Nat Commun. 2018. PMID: 30451839 Free PMC article.
-
Opposing Roles of FANCJ and HLTF Protect Forks and Restrain Replication during Stress.Cell Rep. 2018 Sep 18;24(12):3251-3261. doi: 10.1016/j.celrep.2018.08.065. Cell Rep. 2018. PMID: 30232006 Free PMC article.
-
C-terminal in Sp1-like artificial zinc-finger proteins plays crucial roles in determining their DNA binding affinity.BMC Biotechnol. 2013 Dec 1;13:106. doi: 10.1186/1472-6750-13-106. BMC Biotechnol. 2013. PMID: 24289163 Free PMC article.
-
Polyglutamine Disease Modeling: Epitope Based Screen for Homologous Recombination using CRISPR/Cas9 System.PLoS Curr. 2014 Apr 15;6:ecurrents.hd.0242d2e7ad72225efa72f6964589369a. doi: 10.1371/currents.hd.0242d2e7ad72225efa72f6964589369a. PLoS Curr. 2014. PMID: 24761311 Free PMC article.
-
Upregulating endogenous genes by an RNA-programmable artificial transactivator.Nucleic Acids Res. 2015 Sep 18;43(16):7850-64. doi: 10.1093/nar/gkv682. Epub 2015 Jul 7. Nucleic Acids Res. 2015. PMID: 26152305 Free PMC article.
References
-
- Smogorzewska A., and, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. - PubMed
-
- Hanahan D., and, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Harley CB. Telomerase is not an oncogene. Oncogene. 2002;21:494–502. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

