miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit
- PMID: 23612310
- PMCID: PMC3644088
- DOI: 10.1038/ncomms2742
miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit
Abstract
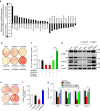
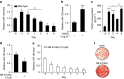
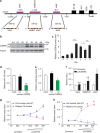
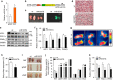
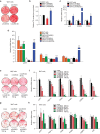
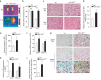
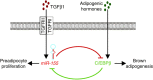
Brown adipocytes are a primary site of energy expenditure and reside not only in classical brown adipose tissue but can also be found in white adipose tissue. Here we show that microRNA 155 is enriched in brown adipose tissue and is highly expressed in proliferating brown preadipocytes but declines after induction of differentiation. Interestingly, microRNA 155 and its target, the adipogenic transcription factor CCAAT/enhancer-binding protein β, form a bistable feedback loop integrating hormonal signals that regulate proliferation or differentiation. Inhibition of microRNA 155 enhances brown adipocyte differentiation and induces a brown adipocyte-like phenotype ('browning') in white adipocytes. Consequently, microRNA 155-deficient mice exhibit increased brown adipose tissue function and 'browning' of white fat tissue. In contrast, transgenic overexpression of microRNA 155 in mice causes a reduction of brown adipose tissue mass and impairment of brown adipose tissue function. These data demonstrate that the bistable loop involving microRNA 155 and CCAAT/enhancer-binding protein β regulates brown lineage commitment, thereby, controlling the development of brown and beige fat cells.
Figures
References
-
- Enerback S.. Human brown adipose tissue. Cell Metab. 11, 248–252 (2010). - PubMed
-
- Nedergaard J. & Cannon B.. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell Metab. 11, 268–272 (2010). - PubMed
-
- van Marken Lichtenbelt W. D. et al.. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360, 1500–1508 (2009). - PubMed
-
- Cinti S.. Transdifferentiation properties of adipocytes in the adipose organ. Am. J. Physiol. Endocrinol. Metab. 297, E977–E986 (2009). - PubMed
-
- Meister G. & Tuschl T.. Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343–349 (2004). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

