Feedback inhibition of deoxy-D-xylulose-5-phosphate synthase regulates the methylerythritol 4-phosphate pathway
- PMID: 23612965
- PMCID: PMC3675625
- DOI: 10.1074/jbc.M113.464636
Feedback inhibition of deoxy-D-xylulose-5-phosphate synthase regulates the methylerythritol 4-phosphate pathway
Abstract
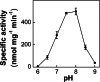
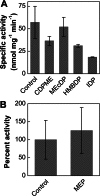
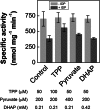
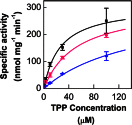
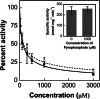
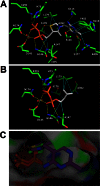
The 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway leads to the biosynthesis of isopentenyl diphosphate (IDP) and dimethylallyl diphosphate (DMADP), the precursors for isoprene and higher isoprenoids. Isoprene has significant effects on atmospheric chemistry, whereas other isoprenoids have diverse roles ranging from various biological processes to applications in commercial uses. Understanding the metabolic regulation of the MEP pathway is important considering the numerous applications of this pathway. The 1-deoxy-D-xylulose-5-phosphate synthase (DXS) enzyme was cloned from Populus trichocarpa, and the recombinant protein (PtDXS) was purified from Escherichia coli. The steady-state kinetic parameters were measured by a coupled enzyme assay. An LC-MS/MS-based assay involving the direct quantification of the end product of the enzymatic reaction, 1-deoxy-D-xylulose 5-phosphate (DXP), was developed. The effect of different metabolites of the MEP pathway on PtDXS activity was tested. PtDXS was inhibited by IDP and DMADP. Both of these metabolites compete with thiamine pyrophosphate for binding with the enzyme. An atomic structural model of PtDXS in complex with thiamine pyrophosphate and Mg(2+) was built by homology modeling and refined by molecular dynamics simulations. The refined structure was used to model the binding of IDP and DMADP and indicated that IDP and DMADP might bind with the enzyme in a manner very similar to the binding of thiamine pyrophosphate. The feedback inhibition of PtDXS by IDP and DMADP constitutes an important mechanism of metabolic regulation of the MEP pathway and indicates that thiamine pyrophosphate-dependent enzymes may often be affected by IDP and DMADP.
Keywords: Deoxyxylulose-5-phosphate Synthase; Enzyme Inhibitors; Enzyme Structure; Isoprene; Isoprenoid; Methylerythritol Pathway; Plant Biochemistry; Thiamine; Thiamine Diphosphate.
Figures


References
-
- Wanke M., Skorupinska-Tudek K., Swiezewska E. (2001) Isoprenoid biosynthesis via 1-deoxy-D-xylulose 5-phosphate/2-C-methyl-D-erythritol 4-phosphate (DOXP/MEP) pathway. Acta Biochim. Pol. 48, 663–672 - PubMed
-
- Hunter W. N. (2007) The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 282, 21573–21577 - PubMed
-
- Phillips M. A., León P., Boronat A., Rodríguez-Concepción M. (2008) The plastidial MEP pathway: unified nomenclature and resources. Trends Plant Sci. 13, 619–623 - PubMed
-
- Hemmerlin A., Hoeffler J. F., Meyer O., Tritsch D., Kagan I. A., Grosdemange-Billiard C., Rohmer M., Bach T. J. (2003) Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright yellow-2 cells. J. Biol. Chem. 278, 26666–26676 - PubMed
-
- Sacchettini J. C., Poulter C. D. (1997) Creating isoprenoid diversity. Science 277, 1788–1789 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

