The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides
- PMID: 23612975
- PMCID: PMC3682527
- DOI: 10.1074/jbc.M112.421685
The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides
Abstract
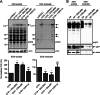
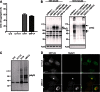
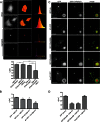
Fragments of proteins containing an expanded polyglutamine (polyQ) tract are thought to initiate aggregation and toxicity in at least nine neurodegenerative diseases, including Huntington's disease. Because proteasomes appear unable to digest the polyQ tract, which can initiate intracellular protein aggregation, preventing polyQ peptide aggregation by chaperones should greatly improve polyQ clearance and prevent aggregate formation. Here we expressed polyQ peptides in cells and show that their intracellular aggregation is prevented by DNAJB6 and DNAJB8, members of the DNAJ (Hsp40) chaperone family. In contrast, HSPA/Hsp70 and DNAJB1, also members of the DNAJ chaperone family, did not prevent peptide-initiated aggregation. Intriguingly, DNAJB6 and DNAJB8 also affected the soluble levels of polyQ peptides, indicating that DNAJB6 and DNAJB8 inhibit polyQ peptide aggregation directly. Together with recent data showing that purified DNAJB6 can suppress fibrillation of polyQ peptides far more efficiently than polyQ expanded protein fragments in vitro, we conclude that the mechanism of DNAJB6 and DNAJB8 is suppression of polyQ protein aggregation by directly binding the polyQ tract.
Keywords: Confocal Microscopy; DNAJB; Huntington's Disease; Molecular Chaperone; Peptides; Polyglutamine Disease.
Figures



Similar articles
-
Co-chaperones DNAJA1 and DNAJB6 are critical for regulation of polyglutamine aggregation.Sci Rep. 2020 May 18;10(1):8130. doi: 10.1038/s41598-020-65046-5. Sci Rep. 2020. PMID: 32424160 Free PMC article.
-
DNAJB6 is a peptide-binding chaperone which can suppress amyloid fibrillation of polyglutamine peptides at substoichiometric molar ratios.Cell Stress Chaperones. 2014 Mar;19(2):227-39. doi: 10.1007/s12192-013-0448-5. Epub 2013 Aug 1. Cell Stress Chaperones. 2014. PMID: 23904097 Free PMC article.
-
Astrocytic expression of the chaperone DNAJB6 results in non-cell autonomous protection in Huntington's disease.Neurobiol Dis. 2019 Apr;124:108-117. doi: 10.1016/j.nbd.2018.10.017. Epub 2018 Nov 5. Neurobiol Dis. 2019. PMID: 30408590
-
Molecular chaperones as modulators of polyglutamine protein aggregation and toxicity.Proc Natl Acad Sci U S A. 2002 Dec 10;99 Suppl 4(Suppl 4):16412-8. doi: 10.1073/pnas.182426899. Epub 2002 Aug 20. Proc Natl Acad Sci U S A. 2002. PMID: 12189209 Free PMC article. Review.
-
Induction of molecular chaperones as a therapeutic strategy for the polyglutamine diseases.Curr Pharm Biotechnol. 2010 Feb;11(2):188-97. doi: 10.2174/138920110790909650. Curr Pharm Biotechnol. 2010. PMID: 20166962 Review.
Cited by
-
DNA methylation study of Huntington's disease and motor progression in patients and in animal models.Nat Commun. 2020 Sep 10;11(1):4529. doi: 10.1038/s41467-020-18255-5. Nat Commun. 2020. PMID: 32913184 Free PMC article.
-
Extracellular chaperone networks and the export of J-domain proteins.J Biol Chem. 2023 Feb;299(2):102840. doi: 10.1016/j.jbc.2022.102840. Epub 2022 Dec 26. J Biol Chem. 2023. PMID: 36581212 Free PMC article. Review.
-
Co-chaperones DNAJA1 and DNAJB6 are critical for regulation of polyglutamine aggregation.Sci Rep. 2020 May 18;10(1):8130. doi: 10.1038/s41598-020-65046-5. Sci Rep. 2020. PMID: 32424160 Free PMC article.
-
Cell and Context-Dependent Effects of the Heat Shock Protein DNAJB6 on Neuronal Survival.Mol Neurobiol. 2016 Oct;53(8):5628-39. doi: 10.1007/s12035-015-9452-3. Epub 2015 Oct 17. Mol Neurobiol. 2016. PMID: 26476842 Free PMC article.
-
Factors affecting protein recovery during Hsp40 affinity profiling.Anal Bioanal Chem. 2024 Aug;416(19):4249-4260. doi: 10.1007/s00216-024-05362-1. Epub 2024 Jun 8. Anal Bioanal Chem. 2024. PMID: 38850318 Free PMC article.
References
-
- Orr H. T., Zoghbi H. Y. (2007) Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30, 575–621 - PubMed
-
- DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 - PubMed
-
- Goti D., Katzen S. M., Mez J., Kurtis N., Kiluk J., Ben-Haïem L., Jenkins N. A., Copeland N. G., Kakizuka A., Sharp A. H., Ross C. A., Mouton P. R., Colomer V. (2004) A mutant ataxin-3 putative-cleavage fragment in brains of Machado-Joseph disease patients and transgenic mice is cytotoxic above a critical concentration. J. Neurosci. 24, 10266–10279 - PMC - PubMed
-
- Lunkes A., Lindenberg K. S., Ben-Haïem L., Weber C., Devys D., Landwehrmeyer G. B., Mandel J. L., Trottier Y. (2002) Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol. Cell 10, 259–269 - PubMed
-
- Cooper J. K., Schilling G., Peters M. F., Herring W. J., Sharp A. H., Kaminsky Z., Masone J., Khan F. A., Delanoy M., Borchelt D. R., Dawson V. L., Dawson T. M., Ross C. A. (1998) Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum. Mol. Genet. 7, 783–790 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

