NHERF2 protein mobility rate is determined by a unique C-terminal domain that is also necessary for its regulation of NHE3 protein in OK cells
- PMID: 23612977
- PMCID: PMC3675628
- DOI: 10.1074/jbc.M113.470799
NHERF2 protein mobility rate is determined by a unique C-terminal domain that is also necessary for its regulation of NHE3 protein in OK cells
Abstract
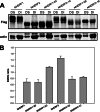
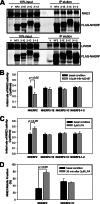
Na(+)/H(+) exchanger regulatory factor (NHERF) proteins are a family of PSD-95/Discs-large/ZO-1 (PDZ)-scaffolding proteins, three of which (NHERFs 1-3) are localized to the brush border in kidney and intestinal epithelial cells. All NHERF proteins are involved in anchoring membrane proteins that contain PDZ recognition motifs to form multiprotein signaling complexes. In contrast to their predicted immobility, NHERF1, NHERF2, and NHERF3 were all shown by fluorescence recovery after photobleaching/confocal microscopy to be surprisingly mobile in the microvilli of the renal proximal tubule OK cell line. Their diffusion coefficients, although different among the three, were all of the same magnitude as that of the transmembrane proteins, suggesting they are all anchored in the microvilli but to different extents. NHERF3 moves faster than NHERF1, and NHERF2 moves the slowest. Several chimeras and mutants of NHERF1 and NHERF2 were made to determine which part of NHERF2 confers the slower mobility rate. Surprisingly, the slower mobility rate of NHERF2 was determined by a unique C-terminal domain, which includes a nonconserved region along with the ezrin, radixin, moesin (ERM) binding domain. Also, this C-terminal domain of NHERF2 determined its greater detergent insolubility and was necessary for the formation of larger multiprotein NHERF2 complexes. In addition, this NHERF2 domain was functionally significant in NHE3 regulation, being necessary for stimulation by lysophosphatidic acid of activity and increased mobility of NHE3, as well as necessary for inhibition of NHE3 activity by calcium ionophore 4-Br-A23187. Thus, multiple functions of NHERF2 require involvement of an additional domain in this protein.
Keywords: Epithelial Cell; Exocytosis; Mobility; Scaffold Proteins; Sodium Proton Exchange; Trafficking.
Figures








References
-
- Lange K. (2011) Fundamental role of microvilli in the main functions of differentiated cells: Outline of an universal regulating and signaling system at the cell periphery. J. Cell. Physiol. 226, 896–927 - PubMed
-
- Brant S. R., Yun C. H., Donowitz M., Tse C. M. (1995) Cloning, tissue distribution, and functional analysis of the human Na+/N+ exchanger isoform, NHE3. Am. J. Physiol. 269, C198–C206 - PubMed
-
- Schultheis P. J., Clarke L. L., Meneton P., Miller M. L., Soleimani M., Gawenis L. R., Riddle T. M., Duffy J. J., Doetschman T., Wang T., Giebisch G., Aronson P. S., Lorenz J. N., Shull G. E. (1998) Renal and intestinal absorptive defects in mice lacking the NHE3 Na+/H+ exchanger. Nat. Genet. 19, 282–285 - PubMed
-
- Donowitz M., Li X. (2007) Regulatory binding partners and complexes of NHE3. Physiol. Rev. 87, 825–872 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

