Helicase loading at chromosomal origins of replication
- PMID: 23613349
- PMCID: PMC3660832
- DOI: 10.1101/cshperspect.a010124
Helicase loading at chromosomal origins of replication
Abstract
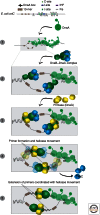
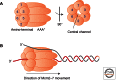
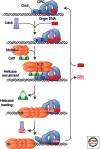
Loading of the replicative DNA helicase at origins of replication is of central importance in DNA replication. As the first of the replication fork proteins assemble at chromosomal origins of replication, the loaded helicase is required for the recruitment of the rest of the replication machinery. In this work, we review the current knowledge of helicase loading at Escherichia coli and eukaryotic origins of replication. In each case, this process requires both an origin recognition protein as well as one or more additional proteins. Comparison of these events shows intriguing similarities that suggest a similar underlying mechanism, as well as critical differences that likely reflect the distinct processes that regulate helicase loading in bacterial and eukaryotic cells.
Figures
References
-
- Abe Y, Jo T, Matsuda Y, Matsunaga C, Katayama T, Ueda T 2007. Structure and function of DnaA N-terminal domains: Specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J Biol Chem 282: 17816–17827 - PubMed
-
- Allen GC, Kornberg A 1991. Fine balance in the regulation of DnaB helicase by DnaC protein in replication in Escherichia coli. J Biol Chem 266: 22096–22101 - PubMed
-
- Aparicio OM, Weinstein DM, Bell SP 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell 91: 59–69 - PubMed
-
- Arai K, Yasuda S, Kornberg A 1981. Mechanism of dnaB protein action. I. Crystallization and properties of dnaB protein, an essential replication protein in Escherichia coli. J Biol Chem 256: 5247–5252 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
