Text mining effectively scores and ranks the literature for improving chemical-gene-disease curation at the comparative toxicogenomics database
- PMID: 23613709
- PMCID: PMC3629079
- DOI: 10.1371/journal.pone.0058201
Text mining effectively scores and ranks the literature for improving chemical-gene-disease curation at the comparative toxicogenomics database
Abstract
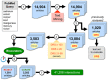
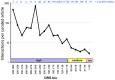
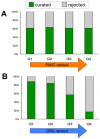
The Comparative Toxicogenomics Database (CTD; http://ctdbase.org/) is a public resource that curates interactions between environmental chemicals and gene products, and their relationships to diseases, as a means of understanding the effects of environmental chemicals on human health. CTD provides a triad of core information in the form of chemical-gene, chemical-disease, and gene-disease interactions that are manually curated from scientific articles. To increase the efficiency, productivity, and data coverage of manual curation, we have leveraged text mining to help rank and prioritize the triaged literature. Here, we describe our text-mining process that computes and assigns each article a document relevancy score (DRS), wherein a high DRS suggests that an article is more likely to be relevant for curation at CTD. We evaluated our process by first text mining a corpus of 14,904 articles triaged for seven heavy metals (cadmium, cobalt, copper, lead, manganese, mercury, and nickel). Based upon initial analysis, a representative subset corpus of 3,583 articles was then selected from the 14,094 articles and sent to five CTD biocurators for review. The resulting curation of these 3,583 articles was analyzed for a variety of parameters, including article relevancy, novel data content, interaction yield rate, mean average precision, and biological and toxicological interpretability. We show that for all measured parameters, the DRS is an effective indicator for scoring and improving the ranking of literature for the curation of chemical-gene-disease information at CTD. Here, we demonstrate how fully incorporating text mining-based DRS scoring into our curation pipeline enhances manual curation by prioritizing more relevant articles, thereby increasing data content, productivity, and efficiency.
Conflict of interest statement
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

