Prospecting for novel plant-derived molecules of Rauvolfia serpentina as inhibitors of Aldose Reductase, a potent drug target for diabetes and its complications
- PMID: 23613832
- PMCID: PMC3629236
- DOI: 10.1371/journal.pone.0061327
Prospecting for novel plant-derived molecules of Rauvolfia serpentina as inhibitors of Aldose Reductase, a potent drug target for diabetes and its complications
Abstract
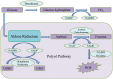
Aldose Reductase (AR) is implicated in the development of secondary complications of diabetes, providing an interesting target for therapeutic intervention. Extracts of Rauvolfia serpentina, a medicinal plant endemic to the Himalayan mountain range, have been known to be effective in alleviating diabetes and its complications. In this study, we aim to prospect for novel plant-derived inhibitors from R. serpentina and to understand structural basis of their interactions. An extensive library of R. serpentina molecules was compiled and computationally screened for inhibitory action against AR. The stability of complexes, with docked leads, was verified using molecular dynamics simulations. Two structurally distinct plant-derived leads were identified as inhibitors: indobine and indobinine. Further, using these two leads as templates, 16 more leads were identified through ligand-based screening of their structural analogs, from a small molecules database. Thus, we obtained plant-derived indole alkaloids, and their structural analogs, as potential AR inhibitors from a manually curated dataset of R. serpentina molecules. Indole alkaloids reported herein, as a novel structural class unreported hitherto, may provide better insights for designing potential AR inhibitors with improved efficacy and fewer side effects.
Conflict of interest statement
Figures











References
-
- Alexiou P, Pegklidou K, Chatzopoulou M, Nicolaou I, Demopoulos V (2009) Aldose reductase enzyme and its implication to major health problems of the 21(st) century. Current Medicinal Chemistry 16: 734–752 doi:10.2174/092986709787458362. - DOI - PubMed
-
- Van Dieren S, Beulens J, Van der Schouw Y, Grobbee D, Neal B (2010) The global burden of diabetes and its complications: an emerging pandemic. European Journal of Cardiovascular Prevention and Rehabilitation 17 Suppl 1S3–8 doi:10.1097/01.hjr.0000368191.86614.5a. - DOI - PubMed
-
- Brownlee M (2001) Biochemistry and molecular cell biology of diabetic complications. Nature 414: 813–820 doi:10.1038/414813a. - DOI - PubMed
-
- Wild S, Roglic G, Green A, Sicree R, King H (2004) Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care 27: 1047–1053 doi:10.2337/diacare.27.5.1047. - DOI - PubMed
-
- Kinoshita J (1990) A thirty year journey in the polyol pathway. Experimental Eye Research 50: 567–573 doi:10.1016/0014-4835(90)90096-D. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

