Widespread divergence of the CEACAM/PSG genes in vertebrates and humans suggests sensitivity to selection
- PMID: 23613906
- PMCID: PMC3628338
- DOI: 10.1371/journal.pone.0061701
Widespread divergence of the CEACAM/PSG genes in vertebrates and humans suggests sensitivity to selection
Abstract
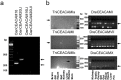
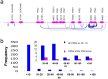
In mammals, carcinoembryonic antigen cell adhesion molecules (CEACAMs) and pregnancy-specific glycoproteins (PSGs) play important roles in the regulation of pathogen transmission, tumorigenesis, insulin signaling turnover, and fetal-maternal interactions. However, how these genes evolved and to what extent they diverged in humans remain to be investigated specifically. Based on syntenic mapping of chordate genomes, we reveal that diverging homologs with a prototypic CEACAM architecture-including an extracellular domain with immunoglobulin variable and constant domain-like regions, and an intracellular domain containing ITAM motif-are present from cartilaginous fish to humans, but are absent in sea lamprey, cephalochordate or urochordate. Interestingly, the CEACAM/PSG gene inventory underwent radical divergence in various vertebrate lineages: from zero in avian species to dozens in therian mammals. In addition, analyses of genetic variations in human populations showed the presence of various types of copy number variations (CNVs) at the CEACAM/PSG locus. These copy number polymorphisms have 3-80% frequency in select populations, and encompass single to more than six PSG genes. Furthermore, we found that CEACAM/PSG genes contain a significantly higher density of nonsynonymous single nucleotide polymorphism (SNP) compared to the chromosome average, and many CEACAM/PSG SNPs exhibit high population differentiation. Taken together, our study suggested that CEACAM/PSG genes have had a more dynamic evolutionary history in vertebrates than previously thought. Given that CEACAM/PSGs play important roles in maternal-fetal interaction and pathogen recognition, these data have laid the groundwork for future analysis of adaptive CEACAM/PSG genotype-phenotypic relationships in normal and complicated pregnancies as well as other etiologies.
Conflict of interest statement
Figures




References
-
- Huang J, Hardy JD, Sun Y, Shively JE (1999) Essential role of biliary glycoprotein (CD66a) in morphogenesis of the human mammary epithelial cell line MCF10F. J Cell Sci 112 (Pt 23): 4193–4205. - PubMed
-
- Leung N, Turbide C, Olson M, Marcus V, Jothy S, et al. (2006) Deletion of the carcinoembryonic antigen-related cell adhesion molecule 1 (Ceacam1) gene contributes to colon tumor progression in a murine model of carcinogenesis. Oncogene 25: 5527–5536. - PubMed
-
- Yokoyama S, Chen CJ, Nguyen T, Shively JE (2007) Role of CEACAM1 isoforms in an in vivo model of mammary morphogenesis: mutational analysis of the cytoplasmic domain of CEACAM1–4S reveals key residues involved in lumen formation. Oncogene 26: 7637–7646. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

