Acidosis activation of the proton-sensing GPR4 receptor stimulates vascular endothelial cell inflammatory responses revealed by transcriptome analysis
- PMID: 23613998
- PMCID: PMC3628782
- DOI: 10.1371/journal.pone.0061991
Acidosis activation of the proton-sensing GPR4 receptor stimulates vascular endothelial cell inflammatory responses revealed by transcriptome analysis
Abstract
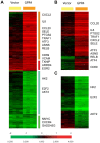
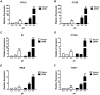
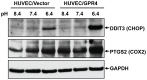
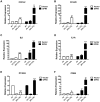
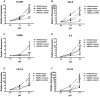
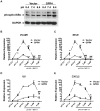
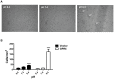
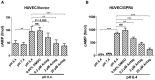
Acidic tissue microenvironment commonly exists in inflammatory diseases, tumors, ischemic organs, sickle cell disease, and many other pathological conditions due to hypoxia, glycolytic cell metabolism and deficient blood perfusion. However, the molecular mechanisms by which cells sense and respond to the acidic microenvironment are not well understood. GPR4 is a proton-sensing receptor expressed in endothelial cells and other cell types. The receptor is fully activated by acidic extracellular pH but exhibits lesser activity at the physiological pH 7.4 and minimal activity at more alkaline pH. To delineate the function and signaling pathways of GPR4 activation by acidosis in endothelial cells, we compared the global gene expression of the acidosis response in primary human umbilical vein endothelial cells (HUVEC) with varying level of GPR4. The results demonstrated that acidosis activation of GPR4 in HUVEC substantially increased the expression of a number of inflammatory genes such as chemokines, cytokines, adhesion molecules, NF-κB pathway genes, and prostaglandin-endoperoxidase synthase 2 (PTGS2 or COX-2) and stress response genes such as ATF3 and DDIT3 (CHOP). Similar GPR4-mediated acidosis induction of the inflammatory genes was also noted in other types of endothelial cells including human lung microvascular endothelial cells and pulmonary artery endothelial cells. Further analyses indicated that the NF-κB pathway was important for the acidosis/GPR4-induced inflammatory gene expression. Moreover, acidosis activation of GPR4 increased the adhesion of HUVEC to U937 monocytic cells under a flow condition. Importantly, treatment with a recently identified GPR4 antagonist significantly reduced the acidosis/GPR4-mediated endothelial cell inflammatory response. Taken together, these results show that activation of GPR4 by acidosis stimulates the expression of a wide range of inflammatory genes in endothelial cells. Such inflammatory response can be suppressed by GPR4 small molecule inhibitors and hold potential therapeutic value.
Conflict of interest statement
Figures
References
-
- Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A (2009) Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 30: 1073–1081. - PubMed
-
- Muller WA (2003) Leukocyte-endothelial-cell interactions in leukocyte transmigration and the inflammatory response. Trends Immunol 24: 327–334. - PubMed
-
- Okpala I (2006) Leukocyte adhesion and the pathophysiology of sickle cell disease. Curr Opin Hematol 13: 40–44. - PubMed
-
- Sughrue ME, Mehra A, Connolly ES Jr, D'Ambrosio AL (2004) Anti-adhesion molecule strategies as potential neuroprotective agents in cerebral ischemia: a critical review of the literature. Inflamm Res 53: 497–508. - PubMed
-
- Castellone RD, Leffler NR, Dong L, Yang LV (2011) Inhibition of tumor cell migration and metastasis by the proton-sensing GPR4 receptor. Cancer Lett. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

