Long-range architecture in a viral RNA genome
- PMID: 23614526
- PMCID: PMC3673720
- DOI: 10.1021/bi4001535
Long-range architecture in a viral RNA genome
Abstract
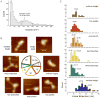
We have developed a model for the secondary structure of the 1058-nucleotide plus-strand RNA genome of the icosahedral satellite tobacco mosaic virus (STMV) using nucleotide-resolution SHAPE chemical probing of the viral RNA isolated from virions and within the virion, perturbation of interactions distant in the primary sequence, and atomic force microscopy. These data are consistent with long-range base pairing interactions and a three-domain genome architecture. The compact domains of the STMV RNA have dimensions of 10-45 nm. Each of the three domains corresponds to a specific functional component of the virus: The central domain corresponds to the coding sequence of the single (capsid) protein encoded by the virus, whereas the 5' and 3' untranslated domains span signals essential for translation and replication, respectively. This three-domain architecture is compatible with interactions between the capsid protein and short RNA helices previously visualized by crystallography. STMV is among the simplest of the icosahedral viruses but, nonetheless, has an RNA genome with a complex higher-order structure that likely reflects high information content and an evolutionary relationship between RNA domain structure and essential replicative functions.
Figures




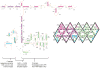
Similar articles
-
Packaged and Free Satellite Tobacco Mosaic Virus (STMV) RNA Genomes Adopt Distinct Conformational States.Biochemistry. 2017 Apr 25;56(16):2175-2183. doi: 10.1021/acs.biochem.6b01166. Epub 2017 Apr 16. Biochemistry. 2017. PMID: 28332826 Free PMC article.
-
Nucleotide sequence and translation of satellite tobacco mosaic virus RNA.Virology. 1989 May;170(1):139-46. doi: 10.1016/0042-6822(89)90361-9. Virology. 1989. PMID: 2718378
-
Double-helical RNA in satellite tobacco mosaic virus.Nature. 1993 Jan 14;361(6408):179-82. doi: 10.1038/361179a0. Nature. 1993. PMID: 8421525
-
Exploring the architecture of viral RNA genomes.Curr Opin Virol. 2015 Jun;12:66-74. doi: 10.1016/j.coviro.2015.03.018. Epub 2015 Apr 14. Curr Opin Virol. 2015. PMID: 25884487 Review.
-
Probing viral genomic structure: alternative viewpoints and alternative structures for satellite tobacco mosaic virus RNA.Biochemistry. 2014 Nov 4;53(43):6728-37. doi: 10.1021/bi501051k. Epub 2014 Oct 24. Biochemistry. 2014. PMID: 25320869 Review.
Cited by
-
Ins and Outs of Multipartite Positive-Strand RNA Plant Viruses: Packaging versus Systemic Spread.Viruses. 2016 Aug 18;8(8):228. doi: 10.3390/v8080228. Viruses. 2016. PMID: 27548199 Free PMC article. Review.
-
Visualizing the global secondary structure of a viral RNA genome with cryo-electron microscopy.RNA. 2015 May;21(5):877-86. doi: 10.1261/rna.047506.114. Epub 2015 Mar 9. RNA. 2015. PMID: 25752599 Free PMC article.
-
Physical and Functional Analysis of Viral RNA Genomes by SHAPE.Annu Rev Virol. 2019 Sep 29;6(1):93-117. doi: 10.1146/annurev-virology-092917-043315. Epub 2019 Jul 23. Annu Rev Virol. 2019. PMID: 31337286 Free PMC article. Review.
-
Progress and outlook in structural biology of large viral RNAs.Virus Res. 2014 Nov 26;193:24-38. doi: 10.1016/j.virusres.2014.06.007. Epub 2014 Jun 21. Virus Res. 2014. PMID: 24956407 Free PMC article. Review.
-
Selective 2'-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis.Nat Protoc. 2015 Nov;10(11):1643-69. doi: 10.1038/nprot.2015.103. Epub 2015 Oct 1. Nat Protoc. 2015. PMID: 26426499 Free PMC article.
References
-
- Koonin EV, Dolja VV. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences, Crit. Rev Biochem Mol Biol. 1993;28:375–430. - PubMed
-
- Schneemann A. The structural and functional role of RNA in icosahedral virus assembly, Annu. Rev Microbiol. 2006;60:51–67. - PubMed
-
- Rao AL. Genome packaging by spherical plant RNA viruses, Annu. Rev Phytopathol. 2006;44:61–87. - PubMed
-
- Hellendoorn K, Mat AW, Gultyaev AP, Pleij CW. Secondary structure model of the coat protein gene of turnip yellow mosaic virus RNA: long, C-rich, single-stranded regions. Virology. 1996;224:43–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

