Evidence for a functionally relevant rocaglamide binding site on the eIF4A-RNA complex
- PMID: 23614532
- PMCID: PMC3796129
- DOI: 10.1021/cb400158t
Evidence for a functionally relevant rocaglamide binding site on the eIF4A-RNA complex
Abstract
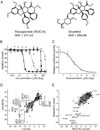
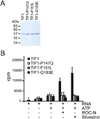
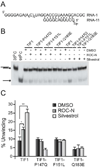
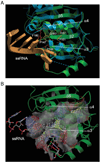
Translation initiation is an emerging target in oncology and neurobiology indications. Naturally derived and synthetic rocaglamide scaffolds have been used to interrogate this pathway; however, there is uncertainty regarding their precise mechanism(s) of action. We exploited the genetic tractability of yeast to define the primary effect of both a natural and a synthetic rocaglamide in a cellular context and characterized the molecular target using biochemical studies and in silico modeling. Chemogenomic profiling and mutagenesis in yeast identified the eIF (eukaryotic Initiation Factor) 4A helicase homologue as the primary molecular target of rocaglamides and defined a discrete set of residues near the RNA binding motif that confer resistance to both compounds. Three of the eIF4A mutations were characterized regarding their functional consequences on activity and response to rocaglamide inhibition. These data support a model whereby rocaglamides stabilize an eIF4A-RNA interaction to either alter the level and/or impair the activity of the eIF4F complex. Furthermore, in silico modeling supports the annotation of a binding pocket delineated by the RNA substrate and the residues identified from our mutagenesis screen. As expected from the high degree of conservation of the eukaryotic translation pathway, these observations are consistent with previous observations in mammalian model systems. Importantly, we demonstrate that the chemically distinct silvestrol and synthetic rocaglamides share a common mechanism of action, which will be critical for optimization of physiologically stable derivatives. Finally, these data confirm the value of the rocaglamide scaffold for exploring the impact of translational modulation on disease.
Figures



References
-
- Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat. Rev. Cancer. 2010;10:254–266. - PubMed
-
- Blagden SP, Willis AE. The biological and therapeutic relevance of mRNA translation in cancer. Nat. Rev. Clin. Oncol. 2011;8:280–291. - PubMed
-
- Lee SK, Cui B, Mehta RR, Kinghorn AD, Pezzuto JM. Cytostatic mechanism and antitumor potential of novel 1H-cyclopenta[b]benzofuran lignans isolated from Aglaia elliptica. Chem. Biol. Interact. 1998;115:215–228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

