Side-chain conformational heterogeneity of intermediates in the Escherichia coli dihydrofolate reductase catalytic cycle
- PMID: 23614825
- PMCID: PMC3796204
- DOI: 10.1021/bi400322e
Side-chain conformational heterogeneity of intermediates in the Escherichia coli dihydrofolate reductase catalytic cycle
Abstract
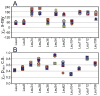
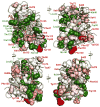
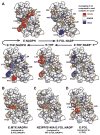
Escherichia coli dihydrofolate reductase (DHFR) provides a paradigm for the integrated study of the role of protein dynamics in enzyme function. Previous studies of backbone and side chain dynamics have yielded unprecedented insights into the mechanism by which DHFR progresses through the structural changes that occur during its catalytic cycle. Here we report a comprehensive study of the χ1 rotamer populations of the aromatic and γ-methyl containing residues for complexes of the catalytic cycle, based on NMR measurement of (3)JCγCO and (3)JCγN coupling constants. We report conformational and dynamic information for eight distinct complexes, where transitions between rotamer wells may occur on a broad picosecond to millisecond time scale. This large volume of (3)J data has allowed us to fit new Karplus parameterizations for aromatic side chains and to select the best available of previously determined parameters for Ile, Thr, and Val. The (3)JCγCO and (3)JCγN coupling constants are found to be extremely sensitive measures of side chain χ1 rotamers and to give important insights into the extent of conformational averaging. For a subset of residues in DHFR, the extent of rotamer averaging is invariant to the nature of the bound ligand, while for other residues the rotamer averaging differs in one or more complexes of the enzymatic cycle. These variable-averaging residues are generally located near the active site, but the phenomenon extends into the adenosine binding domain. For several residues, the rotamer populations in different DHFR complexes appear to depend on whether the complex is in the closed or occluded state, and some residues are exquisitely sensitive to small changes in the nature of the bound ligand.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Bystroff C, Oatley SJ, Kraut J. Crystal structures of Escherichia coli dihydrofolate reductase: the NADP+ holoenzyme and the folate. NADP+ ternary complex Substrate binding and a model for the transition state. Biochemistry. 1990;29:3263–3277. - PubMed
-
- Bystroff C, Kraut J. Crystal structure of unliganded Escherichia coli dihydrofolate reductase. Ligand-induced conformational changes and cooperativity in binding. Biochemistry. 1991;30:2227–2239. - PubMed
-
- Sawaya MR, Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry. 1997;36:586–603. - PubMed
-
- Schnell JR, Dyson HJ, Wright PE. Structure, dynamics and catalytic function of dihydrofolate reductase. Ann Rev Biophys Biomol Struct. 2004;33:119–140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

