Geminivirus protein structure and function
- PMID: 23615043
- PMCID: PMC6638828
- DOI: 10.1111/mpp.12032
Geminivirus protein structure and function
Abstract
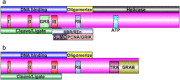
Geminiviruses are a family of plant viruses that cause economically important plant diseases worldwide. These viruses have circular single-stranded DNA genomes and four to eight genes that are expressed from both strands of the double-stranded DNA replicative intermediate. The transcription of these genes occurs under the control of two bidirectional promoters and one monodirectional promoter. The viral proteins function to facilitate virus replication, virus movement, the assembly of virus-specific nucleoprotein particles, vector transmission and to counteract plant host defence responses. Recent research findings have provided new insights into the structure and function of these proteins and have identified numerous host interacting partners. Most of the viral proteins have been shown to be multifunctional, participating in multiple events during the infection cycle and have, indeed, evolved coordinated interactions with host proteins to ensure a successful infection. Here, an up-to-date review of viral protein structure and function is presented, and some areas requiring further research are identified.
© 2013 BSPP AND JOHN WILEY & SONS LTD.
Figures






References
-
- Avvakumov, N. , Sahbegovic, M. , Zhang, Z. , Shuen, M. and Mymryk, J.S. (2002) Analysis of DNA binding by the adenovirus type 5 E1A oncoprotein. J. Gen. Virol. 83, 517–524. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

