Phosphorylation of serine 1137/1138 of mouse insulin receptor substrate (IRS) 2 regulates cAMP-dependent binding to 14-3-3 proteins and IRS2 protein degradation
- PMID: 23615913
- PMCID: PMC3675577
- DOI: 10.1074/jbc.M113.474593
Phosphorylation of serine 1137/1138 of mouse insulin receptor substrate (IRS) 2 regulates cAMP-dependent binding to 14-3-3 proteins and IRS2 protein degradation
Abstract
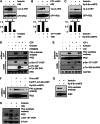
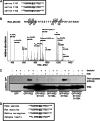
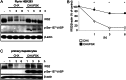
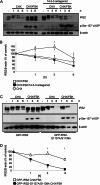
Insulin receptor substrate (IRS) 2 as intermediate docking platform transduces the insulin/IGF-1 (insulin like growth factor 1) signal to intracellular effector molecules that regulate glucose homeostasis, β-cell growth, and survival. Previously, IRS2 has been identified as a 14-3-3 interaction protein. 14-3-3 proteins can bind their target proteins via phosphorylated serine/threonine residues located within distinct motifs. In this study the binding of 14-3-3 to IRS2 upon stimulation with forskolin or the cAMP analog 8-(4-chlorophenylthio)-cAMP was demonstrated in HEK293 cells. Binding was reduced with PKA inhibitors H89 or Rp-8-Br-cAMPS. Phosphorylation of IRS2 on PKA consensus motifs was induced by forskolin and the PKA activator N(6)-Phe-cAMP and prevented by both PKA inhibitors. The amino acid region after position 952 on IRS2 was identified as the 14-3-3 binding region by GST-14-3-3 pulldown assays. Mass spectrometric analysis revealed serine 1137 and serine 1138 as cAMP-dependent, potential PKA phosphorylation sites. Mutation of serine 1137/1138 to alanine strongly reduced the cAMP-dependent 14-3-3 binding. Application of cycloheximide revealed that forskolin enhanced IRS2 protein stability in HEK293 cells stably expressing IRS2 as well as in primary hepatocytes. Stimulation with forskolin did not increase protein stability either in the presence of a 14-3-3 antagonist or in the double 1137/1138 alanine mutant. Thus the reduced IRS2 protein degradation was dependent on the interaction with 14-3-3 proteins and the presence of serine 1137/1138. We present serine 1137/1138 as novel cAMP-dependent phosphorylation sites on IRS2 and show their importance in 14-3-3 binding and IRS2 protein stability.
Keywords: 14-3-3; Cyclic AMP (cAMP); IRS2; Insulin; Phosphorylation; Protein Degradation; Protein Kinase A (PKA).
Figures


Similar articles
-
Identification of the amino acids 300-600 of IRS-2 as 14-3-3 binding region with the importance of IGF-1/insulin-regulated phosphorylation of Ser-573.PLoS One. 2012;7(8):e43296. doi: 10.1371/journal.pone.0043296. Epub 2012 Aug 17. PLoS One. 2012. PMID: 22912850 Free PMC article.
-
Plk1 phosphorylation of IRS2 prevents premature mitotic exit via AKT inactivation.Biochemistry. 2015 Apr 21;54(15):2473-80. doi: 10.1021/acs.biochem.5b00016. Epub 2015 Apr 6. Biochemistry. 2015. PMID: 25830382 Free PMC article.
-
Mechanism of protein kinase B activation by cyclic AMP-dependent protein kinase.Mol Cell Biol. 1999 Jul;19(7):4989-5000. doi: 10.1128/MCB.19.7.4989. Mol Cell Biol. 1999. PMID: 10373549 Free PMC article.
-
Insulin induces IRS2-dependent and GRK2-mediated β2AR internalization to attenuate βAR signaling in cardiomyocytes.Cell Signal. 2015 Mar;27(3):707-15. doi: 10.1016/j.cellsig.2014.11.018. Epub 2014 Nov 25. Cell Signal. 2015. PMID: 25460042 Free PMC article.
-
Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2.Diabetologia. 2012 Oct;55(10):2565-2582. doi: 10.1007/s00125-012-2644-8. Epub 2012 Aug 8. Diabetologia. 2012. PMID: 22869320 Free PMC article. Review.
Cited by
-
Prolyl Carboxypeptidase Maintains Receptor Tyrosine Kinase Signaling and Is a Potential Therapeutic Target in Triple Negative Breast Cancer.Cancers (Basel). 2022 Jan 31;14(3):739. doi: 10.3390/cancers14030739. Cancers (Basel). 2022. PMID: 35159006 Free PMC article.
-
The dynamic and stress-adaptive signaling hub of 14-3-3: emerging mechanisms of regulation and context-dependent protein-protein interactions.Oncogene. 2018 Oct;37(42):5587-5604. doi: 10.1038/s41388-018-0348-3. Epub 2018 Jun 18. Oncogene. 2018. PMID: 29915393 Free PMC article. Review.
-
14-3-3-β and -{varepsilon} contribute to activation of the osmoprotective transcription factor NFAT5 by increasing its protein abundance and its transactivating activity.Physiol Rep. 2014 Apr 22;2(4):e12000. doi: 10.14814/phy2.12000. Print 2014. Physiol Rep. 2014. PMID: 24771694 Free PMC article.
-
Is 14-3-3 the Combination to Unlock New Pathways to Improve Metabolic Homeostasis and β-Cell Function?Diabetes. 2023 Aug 1;72(8):1045-1054. doi: 10.2337/db23-0094. Diabetes. 2023. PMID: 37471599 Free PMC article. Review.
-
14-3-3ζ coordinates adipogenesis of visceral fat.Nat Commun. 2015 Jul 29;6:7671. doi: 10.1038/ncomms8671. Nat Commun. 2015. PMID: 26220403 Free PMC article.
References
-
- Kubota N., Tobe K., Terauchi Y., Eto K., Yamauchi T., Suzuki R., Tsubamoto Y., Komeda K., Nakano R., Miki H., Satoh S., Sekihara H., Sciacchitano S., Lesniak M., Aizawa S., Nagai R., Kimura S., Akanuma Y., Taylor S. I., Kadowaki T. (2000) Disruption of insulin receptor substrate 2 causes type 2 diabetes because of liver insulin resistance and lack of compensatory beta-cell hyperplasia. Diabetes 49, 1880–1889 - PubMed
-
- Kubota N., Kubota T., Itoh S., Kumagai H., Kozono H., Takamoto I., Mineyama T., Ogata H., Tokuyama K., Ohsugi M., Sasako T., Moroi M., Sugi K., Kakuta S., Iwakura Y., Noda T., Ohnishi S., Nagai R., Tobe K., Terauchi Y., Ueki K., Kadowaki T. (2008) Dynamic functional relay between insulin receptor substrate 1 and 2 in hepatic insulin signaling during fasting and feeding. Cell Metab. 8, 49–64 - PubMed
-
- Withers D. J., Gutierrez J. S., Towery H., Burks D. J., Ren J. M., Previs S., Zhang Y., Bernal D., Pons S., Shulman G. I., Bonner-Weir S., White M. F. (1998) Disruption of IRS-2 causes type 2 diabetes in mice. Nature 391, 900–904 - PubMed
-
- Schuppin G. T., Pons S., Hügl S., Aiello L. P., King G. L., White M., Rhodes C. J. (1998) A specific increased expression of insulin receptor substrate 2 in pancreatic beta-cell lines is involved in mediating serum-stimulated beta-cell growth. Diabetes 47, 1074–1085 - PubMed
-
- Hennige A. M., Burks D. J., Ozcan U., Kulkarni R. N., Ye J., Park S., Schubert M., Fisher T. L., Dow M. A., Leshan R., Zakaria M., Mossa-Basha M., White M. F. (2003) Up-regulation of insulin receptor substrate-2 in pancreatic beta cells prevents diabetes. J. Clin. Invest. 112, 1521–1532 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

