IRF-5 and NF-κB p50 co-regulate IFN-β and IL-6 expression in TLR9-stimulated human plasmacytoid dendritic cells
- PMID: 23616277
- PMCID: PMC6389267
- DOI: 10.1002/eji.201242792
IRF-5 and NF-κB p50 co-regulate IFN-β and IL-6 expression in TLR9-stimulated human plasmacytoid dendritic cells
Abstract
Synthetic oligonucleotides (ODN) expressing CpG motifs mimic the ability of bacterial DNA to trigger the innate immune system via TLR9. Plasmacytoid dendritic cells (pDCs) make a critical contribution to the ensuing immune response. This work examines the induction of antiviral (IFN-β) and pro-inflammatory (IL-6) cytokines by CpG-stimulated human pDCs and the human CAL-1 pDC cell line. Results show that interferon regulatory factor-5 (IRF-5) and NF-κB p50 are key co-regulators of IFN-β and IL-6 expression following TLR9-mediated activation of human pDCs. The nuclear accumulation of IRF-1 was also observed, but this was a late event that was dependant on type 1 IFN and unrelated to the initiation of gene expression. IRF-8 was identified as a novel negative regulator of gene activation in CpG-stimulated pDCs. As variants of IRF-5 and IRF-8 were recently found to correlate with susceptibility to certain autoimmune diseases, these findings are relevant to our understanding of the pharmacologic effects of "K" ODN and the role of TLR9 ligation under physiologic, pathologic, and therapeutic conditions.
Keywords: CpG oligonucleotide; Dendritic cell; IRF-5; NF-κB; TLR9.
Published 2013. This is a US Government work and is in the public domain in the USA. Published by WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Conflict of interest:
Dr. Dennis Klinman and members of his lab are co-inventors on a number of patents concerning CpG ODN and their use. All rights to these patents have been assigned to the Federal government.
Figures
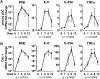


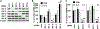

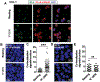

Comment in
-
IRF5, IRF8, and IRF7 in human pDCs - the good, the bad, and the insignificant?Eur J Immunol. 2013 Jul;43(7):1693-7. doi: 10.1002/eji.201343739. Eur J Immunol. 2013. PMID: 23828296
References
-
- Kawai T and Akira S, The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol 2010. 11: 373–384. - PubMed
-
- Jarrossay D, Napolitani G, Colonna M, Sallusto F and Lanzavecchia A, Specialization and complementarity in microbial molecule recognition by human myeloid and plasmacytoid dendritic cells. Eur. J. Immunol 2001. 31: 3388–3393. - PubMed
-
- Haining WN, Davies J, Kanzler H, Drury L, Brenn T, Evans J, Angelosanto J et al., CpG oligodeoxynucleotides alter lymphocyte and dendritic cell trafficking in humans. Clin. Cancer Res. 2008. 14: 5626–5634. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

