A conserved gammaherpesvirus protein kinase targets histone deacetylases 1 and 2 to facilitate viral replication in primary macrophages
- PMID: 23616648
- PMCID: PMC3700300
- DOI: 10.1128/JVI.02713-12
A conserved gammaherpesvirus protein kinase targets histone deacetylases 1 and 2 to facilitate viral replication in primary macrophages
Erratum in
-
Correction for Mounce et al., "A Conserved Gammaherpesvirus Protein Kinase Targets Histone Deacetylases 1 and 2 To Facilitate Viral Replication in Primary Macrophages".J Virol. 2017 Dec 14;92(1):e01601-17. doi: 10.1128/JVI.01601-17. Print 2018 Jan 1. J Virol. 2017. PMID: 29242252 Free PMC article. No abstract available.
Abstract
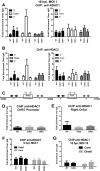
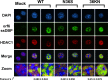
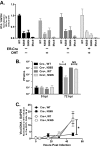
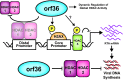
Gammaherpesviruses are ubiquitious pathogens that establish lifelong infection and are associated with several malignancies. All gammaherpesviruses encode a conserved protein kinase that facilitates viral replication and chronic infection and thus represents an attractive therapeutic target. In this study, we identify a novel function of gammaherpesvirus protein kinase as a regulator of class I histone deacetylases (HDAC). Mouse gammaherpesvirus 68 (MHV68)-encoded protein kinase orf36 interacted with HDAC1 and 2 and prevented association of these HDACs with the viral promoter driving expression of RTA, a critical immediate early transcriptional activator. Furthermore, the ability to interact with HDAC1 and 2 was not limited to the MHV68 orf36, as BGLF4, a related viral protein kinase encoded by Epstein-Barr virus, interacted with HDAC1 in vitro. Importantly, targeting of HDAC1 and 2 by orf36 was independent of the kinase's enzymatic activity. Additionally, orf36 expression, but not its enzymatic activity, induced changes in the global deacetylase activity observed in infected primary macrophages. Combined deficiency of HDAC1 and 2 rescued attenuated replication and viral DNA synthesis of the orf36 null MHV68 mutant, indicating that the regulation of HDAC1 and 2 by orf36 was relevant for viral replication. Understanding the mechanism by which orf36 facilitates viral replication, including through HDAC targeting, will facilitate the development of improved therapeutics against gammaherpesvirus kinases.
Figures
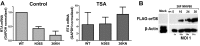



Similar articles
-
Primary macrophages rely on histone deacetylase 1 and 2 expression to induce type I interferon in response to gammaherpesvirus infection.J Virol. 2014 Feb;88(4):2268-78. doi: 10.1128/JVI.03278-13. Epub 2013 Dec 11. J Virol. 2014. PMID: 24335310 Free PMC article.
-
Gammaherpesvirus gene expression and DNA synthesis are facilitated by viral protein kinase and histone variant H2AX.Virology. 2011 Nov 25;420(2):73-81. doi: 10.1016/j.virol.2011.08.019. Epub 2011 Sep 22. Virology. 2011. PMID: 21943826 Free PMC article.
-
Conserved Herpesvirus Kinase ORF36 Activates B2 Retrotransposons during Murine Gammaherpesvirus Infection.J Virol. 2020 Jul 1;94(14):e00262-20. doi: 10.1128/JVI.00262-20. Print 2020 Jul 1. J Virol. 2020. PMID: 32404524 Free PMC article.
-
Hijacking the DNA damage response to enhance viral replication: gamma-herpesvirus 68 orf36 phosphorylates histone H2AX.Mol Cell. 2007 Jul 20;27(2):178-179. doi: 10.1016/j.molcel.2007.07.005. Mol Cell. 2007. PMID: 17643367 Free PMC article. Review.
-
Histone deacetylases in herpesvirus replication and virus-stimulated host defense.Viruses. 2013 Jun 27;5(7):1607-32. doi: 10.3390/v5071607. Viruses. 2013. PMID: 23807710 Free PMC article. Review.
Cited by
-
STAT6 degradation and ubiquitylated TRIML2 are essential for activation of human oncogenic herpesvirus.PLoS Pathog. 2018 Dec 10;14(12):e1007416. doi: 10.1371/journal.ppat.1007416. eCollection 2018 Dec. PLoS Pathog. 2018. PMID: 30532138 Free PMC article.
-
A locus encompassing the Epstein-Barr virus bglf4 kinase regulates expression of genes encoding viral structural proteins.PLoS Pathog. 2014 Aug 28;10(8):e1004307. doi: 10.1371/journal.ppat.1004307. eCollection 2014 Aug. PLoS Pathog. 2014. PMID: 25166506 Free PMC article.
-
Marek's disease virus US3 protein kinase phosphorylates chicken HDAC 1 and 2 and regulates viral replication and pathogenesis.PLoS Pathog. 2021 Feb 17;17(2):e1009307. doi: 10.1371/journal.ppat.1009307. eCollection 2021 Feb. PLoS Pathog. 2021. PMID: 33596269 Free PMC article.
-
Hsp90 inhibitor 17-DMAG decreases expression of conserved herpesvirus protein kinases and reduces virus production in Epstein-Barr virus-infected cells.J Virol. 2013 Sep;87(18):10126-38. doi: 10.1128/JVI.01671-13. Epub 2013 Jul 10. J Virol. 2013. PMID: 23843639 Free PMC article.
-
Histone H3K14 hypoacetylation and H3K27 hypermethylation along with HDAC1 up-regulation and KDM6B down-regulation are associated with active pulmonary tuberculosis disease.Am J Transl Res. 2017 Apr 15;9(4):1943-1955. eCollection 2017. Am J Transl Res. 2017. PMID: 28469799 Free PMC article.
References
-
- Sakamoto S, Potla R, Larner AC. 2004. Histone deacetylase activity is required to recruit RNA polymerase II to the promoters of selected interferon-stimulated early response genes. J. Biol. Chem. 279:40362–40367 - PubMed
-
- Peng L, Seto E. 2011. Deacetylation of nonhistone proteins by HDACs and the implications in cancer. Handb. Exp. Pharmacol. 206:39–56 - PubMed
-
- Witt O, Deubzer HE, Milde T, Oehme I. 2009. HDAC family: what are the cancer relevant targets? Cancer Lett. 277:8–21 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

