Identification of multiple novel viruses, including a parvovirus and a hepevirus, in feces of red foxes
- PMID: 23616657
- PMCID: PMC3700315
- DOI: 10.1128/JVI.00568-13
Identification of multiple novel viruses, including a parvovirus and a hepevirus, in feces of red foxes
Abstract
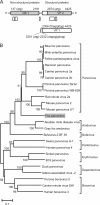
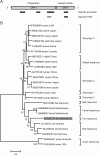
Red foxes (Vulpes vulpes) are the most widespread members of the order of Carnivora. Since they often live in (peri)urban areas, they are a potential reservoir of viruses that transmit from wildlife to humans or domestic animals. Here we evaluated the fecal viral microbiome of 13 red foxes by random PCR in combination with next-generation sequencing. Various novel viruses, including a parvovirus, bocavirus, adeno-associated virus, hepevirus, astroviruses, and picobirnaviruses, were identified.
Figures

References
-
- World Health Organization 2012. Outbreak news. Ebola, Democratic Republic of Congo—update. Wkly. Epidemiol. Rec. 87:357 http://www.who.int/wer - PubMed
-
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367:1814–1820 - PubMed
-
- Mahalingam S, Herrero LJ, Playford EG, Spann K, Herring B, Rolph MS, Middleton D, McCall B, Field H, Wang LF. 2012. Hendra virus: an emerging paramyxovirus in Australia. Lancet Infect. Dis. 12:799–807 - PubMed
-
- Kuiken T, Leighton FA, Fouchier RA, LeDuc JW, Peiris JS, Schudel A, Stohr K, Osterhaus AD. 2005. Public health. Pathogen surveillance in animals. Science 309:1680–1681 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

