Quantification of N-acetylcysteamine activated methylmalonate incorporation into polyketide biosynthesis
- PMID: 23616811
- PMCID: PMC3628877
- DOI: 10.3762/bjoc.9.75
Quantification of N-acetylcysteamine activated methylmalonate incorporation into polyketide biosynthesis
Abstract
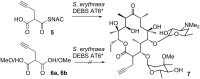
Polyketides are biosynthesized through consecutive decarboxylative Claisen condensations between a carboxylic acid and differently substituted malonic acid thioesters, both tethered to the giant polyketide synthase enzymes. Individual malonic acid derivatives are typically required to be activated as coenzyme A-thioesters prior to their enzyme-catalyzed transfer onto the polyketide synthase. Control over the selection of malonic acid building blocks promises great potential for the experimental alteration of polyketide structure and bioactivity. One requirement for this endeavor is the supplementation of the bacterial polyketide fermentation system with tailored synthetic thioester-activated malonates. The membrane permeable N-acetylcysteamine has been proposed as a coenzyme A-mimic for this purpose. Here, the incorporation efficiency into different polyketides of N-acetylcysteamine activated methylmalonate is studied and quantified, showing a surprisingly high and transferable activity of these polyketide synthase substrate analogues in vivo.
Keywords: biosynthesis; coenzyme A; malonic acid; polyketide; polyketide synthase.
Figures

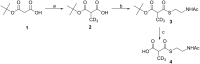
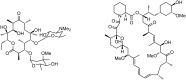

Similar articles
-
Predicted incorporation of non-native substrates by a polyketide synthase yields bioactive natural product derivatives.Chembiochem. 2014 Sep 5;15(13):1991-7. doi: 10.1002/cbic.201402206. Epub 2014 Jul 15. Chembiochem. 2014. PMID: 25044264
-
Flexible enzymatic activation of artificial polyketide extender units by Streptomyces cinnamonensis into the monensin biosynthetic pathway.Lett Appl Microbiol. 2018 Sep;67(3):226-234. doi: 10.1111/lam.13039. Epub 2018 Jul 18. Lett Appl Microbiol. 2018. PMID: 29927502
-
Biomimetic Thioesters as Probes for Enzymatic Assembly Lines: Synthesis, Applications, and Challenges.Cell Chem Biol. 2016 Oct 20;23(10):1179-1192. doi: 10.1016/j.chembiol.2016.08.014. Epub 2016 Sep 29. Cell Chem Biol. 2016. PMID: 27693058 Review.
-
Enzyme-directed mutasynthesis: a combined experimental and theoretical approach to substrate recognition of a polyketide synthase.ACS Chem Biol. 2013 Feb 15;8(2):443-50. doi: 10.1021/cb300505w. Epub 2012 Nov 26. ACS Chem Biol. 2013. PMID: 23181268
-
Structure and function of polyketide biosynthetic enzymes: various strategies for production of structurally diverse polyketides.Biosci Biotechnol Biochem. 2017 Dec;81(12):2227-2236. doi: 10.1080/09168451.2017.1391687. Epub 2017 Nov 1. Biosci Biotechnol Biochem. 2017. PMID: 29090652 Review.
Cited by
-
Acyltransferases as Tools for Polyketide Synthase Engineering.Antibiotics (Basel). 2018 Jul 18;7(3):62. doi: 10.3390/antibiotics7030062. Antibiotics (Basel). 2018. PMID: 30022008 Free PMC article. Review.
-
Engineering the acyltransferase substrate specificity of assembly line polyketide synthases.J R Soc Interface. 2013 May 29;10(85):20130297. doi: 10.1098/rsif.2013.0297. Print 2013 Aug 6. J R Soc Interface. 2013. PMID: 23720536 Free PMC article. Review.
-
Structural Insight of KSIII (β-Ketoacyl-ACP Synthase)-like Acyltransferase ChlB3 in the Biosynthesis of Chlorothricin.Molecules. 2022 Sep 28;27(19):6405. doi: 10.3390/molecules27196405. Molecules. 2022. PMID: 36234941 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
