TRIAD: The Translational Research Informatics and Data Management Grid
- PMID: 23616879
- PMCID: PMC3631927
- DOI: 10.4338/ACI-2011-02-RA-0014
TRIAD: The Translational Research Informatics and Data Management Grid
Abstract
Objective: Multi-disciplinary and multi-site biomedical research programs frequently require infrastructures capable of enabling the collection, management, analysis, and dissemination of heterogeneous, multi-dimensional, and distributed data and knowledge collections spanning organizational boundaries. We report on the design and initial deployment of an extensible biomedical informatics platform that is intended to address such requirements.
Methods: A common approach to distributed data, information, and knowledge management needs in the healthcare and life science settings is the deployment and use of a service-oriented architecture (SOA). Such SOA technologies provide for strongly-typed, semantically annotated, and stateful data and analytical services that can be combined into data and knowledge integration and analysis "pipelines." Using this overall design pattern, we have implemented and evaluated an extensible SOA platform for clinical and translational science applications known as the Translational Research Informatics and Data-management grid (TRIAD). TRIAD is a derivative and extension of the caGrid middleware and has an emphasis on supporting agile "working interoperability" between data, information, and knowledge resources.
Results: Based upon initial verification and validation studies conducted in the context of a collection of driving clinical and translational research problems, we have been able to demonstrate that TRIAD achieves agile "working interoperability" between distributed data and knowledge sources.
Conclusion: Informed by our initial verification and validation studies, we believe TRIAD provides an example instance of a lightweight and readily adoptable approach to the use of SOA technologies in the clinical and translational research setting. Furthermore, our initial use cases illustrate the importance and efficacy of enabling "working interoperability" in heterogeneous biomedical environments.
Keywords: Clinical research informatics; data access; data analysis; data integration; socio-organizational issues; standards; workflow.
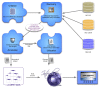
Figures



Similar articles
-
Adoption and Adaptation of caGrid for CTSA.Summit Transl Bioinform. 2009 Mar 1;2009:44-8. Summit Transl Bioinform. 2009. PMID: 21347169 Free PMC article.
-
A roadmap for caGrid, an enterprise Grid architecture for biomedical research.Stud Health Technol Inform. 2008;138:224-37. Stud Health Technol Inform. 2008. PMID: 18560123 Free PMC article.
-
caGrid: design and implementation of the core architecture of the cancer biomedical informatics grid.Bioinformatics. 2006 Aug 1;22(15):1910-6. doi: 10.1093/bioinformatics/btl272. Epub 2006 Jun 9. Bioinformatics. 2006. PMID: 16766552
-
Translational informatics: enabling high-throughput research paradigms.Physiol Genomics. 2009 Nov 6;39(3):131-40. doi: 10.1152/physiolgenomics.00050.2009. Epub 2009 Sep 8. Physiol Genomics. 2009. PMID: 19737991 Free PMC article. Review.
-
Building essential biodiversity variables (EBVs) of species distribution and abundance at a global scale.Biol Rev Camb Philos Soc. 2018 Feb;93(1):600-625. doi: 10.1111/brv.12359. Epub 2017 Aug 2. Biol Rev Camb Philos Soc. 2018. PMID: 28766908 Review.
Cited by
-
A survey of informatics platforms that enable distributed comparative effectiveness research using multi-institutional heterogenous clinical data.Med Care. 2012 Jul;50 Suppl(Suppl):S49-59. doi: 10.1097/MLR.0b013e318259c02b. Med Care. 2012. PMID: 22692259 Free PMC article.
-
Using patient lists to add value to integrated data repositories.J Biomed Inform. 2014 Dec;52:72-7. doi: 10.1016/j.jbi.2014.02.010. Epub 2014 Feb 15. J Biomed Inform. 2014. PMID: 24534444 Free PMC article.
-
A Modular Architecture for Electronic Health Record-Driven Phenotyping.AMIA Jt Summits Transl Sci Proc. 2015 Mar 25;2015:147-51. eCollection 2015. AMIA Jt Summits Transl Sci Proc. 2015. PMID: 26306258 Free PMC article.
-
A system to build distributed multivariate models and manage disparate data sharing policies: implementation in the scalable national network for effectiveness research.J Am Med Inform Assoc. 2015 Nov;22(6):1187-95. doi: 10.1093/jamia/ocv017. Epub 2015 Jul 3. J Am Med Inform Assoc. 2015. PMID: 26142423 Free PMC article.
-
Integration and Visualization of Translational Medicine Data for Better Understanding of Human Diseases.Big Data. 2016 Jun;4(2):97-108. doi: 10.1089/big.2015.0057. Big Data. 2016. PMID: 27441714 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

