Analysis of the factor XI variant Arg184Gly suggests a structural basis for factor IX binding to factor XIa
- PMID: 23617568
- PMCID: PMC4158700
- DOI: 10.1111/jth.12275
Analysis of the factor XI variant Arg184Gly suggests a structural basis for factor IX binding to factor XIa
Abstract
Background: A patient with factor XI (FXI) deficiency was reported with an Arg184Gly substitution in the FXI A3 domain. The A3 domain contains an exosite required for binding of FIX to activated FXI (FXIa).
Objective: To test the effects of the Arg184Gly substitution on FIX activation, and to characterize the FIX-binding site on FXIa.
Methods: Recombinant FXIa and FIX variants were used to identify residues involved in FIX activation by FXIa. Analysis of the FXI structure was used to identify potential FIX-binding sites.
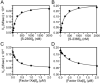
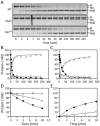
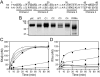
Results: The Km for FIX activation by FXIa-Gly184 was approximately three-fold higher than for FXIa, suggesting that Arg184 is part of the exosite. Arg184 and the adjacent residues, Ile183 and Asp185, contribute to charged and hydrophobic areas that are not present in the FXI homolog prekallikrein (PK). Replacing residues 183-185 with alanine abolished exosite activity, similarly to replacement of the entire A3 domain with the A3 domain from PK (FXIa/PKA3). Reintroducing FXI residues 183-185 into FXIa/PKA3 partially restored the exosite, and replacing residues 183-185 and 260-264 completely restored exosite function. FIX in which the Ω-loop (residues 4-11) was replaced with the FVII Ω-loop was activated poorly by FXIa, suggesting that the FIX Ω-loop binds to FXIa.
Conclusions: The results support a model in which the Ω-loop of FIX binds to an area on FXIa composed of residues from the N-terminus and C-terminus of the A3 domain. These residues are buried in zymogen FXI, and must be exposed upon conversion to FXIa to permit FIX binding.
Keywords: blood coagulation; factor IX; factor XI; factor XIa; proteolysis.
© 2013 International Society on Thrombosis and Haemostasis.
Conflict of interest statement
Figures




Similar articles
-
Hydrogen-deuterium exchange mass spectrometry highlights conformational changes induced by factor XI activation and binding of factor IX to factor XIa.J Thromb Haemost. 2019 Dec;17(12):2047-2055. doi: 10.1111/jth.14632. Epub 2019 Oct 7. J Thromb Haemost. 2019. PMID: 31519061 Free PMC article.
-
The mechanism underlying activation of factor IX by factor XIa.Thromb Res. 2014 May;133 Suppl 1(0 1):S48-51. doi: 10.1016/j.thromres.2014.03.020. Thromb Res. 2014. PMID: 24759143 Free PMC article. Review.
-
A sequential mechanism for exosite-mediated factor IX activation by factor XIa.J Biol Chem. 2012 Nov 2;287(45):38200-9. doi: 10.1074/jbc.M112.376343. Epub 2012 Sep 7. J Biol Chem. 2012. PMID: 22961984 Free PMC article.
-
Productive recognition of factor IX by factor XIa exosites requires disulfide linkage between heavy and light chains of factor XIa.J Biol Chem. 2012 Feb 24;287(9):6187-95. doi: 10.1074/jbc.M111.291989. Epub 2011 Dec 29. J Biol Chem. 2012. PMID: 22207756 Free PMC article.
-
Structure and function of factor XI.Blood. 2010 Apr 1;115(13):2569-77. doi: 10.1182/blood-2009-09-199182. Epub 2010 Jan 28. Blood. 2010. PMID: 20110423 Free PMC article. Review.
Cited by
-
The evolution of factor XI and the kallikrein-kinin system.Blood Adv. 2020 Dec 22;4(24):6135-6147. doi: 10.1182/bloodadvances.2020002456. Blood Adv. 2020. PMID: 33351111 Free PMC article.
-
Evidence for factor IX-independent roles for factor XIa in blood coagulation.J Thromb Haemost. 2013 Dec;11(12):2118-27. doi: 10.1111/jth.12435. J Thromb Haemost. 2013. PMID: 24152424 Free PMC article.
-
Factor XI as a target for antithrombotic therapy.Drug Discov Today. 2014 Sep;19(9):1454-8. doi: 10.1016/j.drudis.2014.05.018. Epub 2014 Jun 2. Drug Discov Today. 2014. PMID: 24886766 Free PMC article. Review.
-
Nanobodies against factor XI apple 3 domain inhibit binding of factor IX and reveal a novel binding site for high molecular weight kininogen.J Thromb Haemost. 2022 Nov;20(11):2538-2549. doi: 10.1111/jth.15815. Epub 2022 Jul 20. J Thromb Haemost. 2022. PMID: 35815349 Free PMC article.
-
Skeletal muscle myosin promotes coagulation by binding factor XI via its A3 domain and enhancing thrombin-induced factor XI activation.J Biol Chem. 2022 Feb;298(2):101567. doi: 10.1016/j.jbc.2022.101567. Epub 2022 Jan 7. J Biol Chem. 2022. PMID: 35007530 Free PMC article.
References
-
- Brummel-Ziedens K, Mann KG. Hematology: basic principles and practice. 6th. Churchill Livingstone-Elsevier; Philadelphia: 2012. Molecular basis of blood coagulation; pp. 1821–41.
-
- Vadivel K, Schmidt AE, Marder VJ, Krishnaswamy S, Bajaj SP. Hemostasis and Thrombosis: basic principles and clinical practice. 6th. Lippincott, Williams & Wilkins; Philadelphia: 2012. Structure and function of vitamin K-dependent coagulation and anticoagulation proteins; pp. 208–32.
-
- Gailani D, Renné T, Emsley J. Factor XI and the Plasma contact system. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; NY, NY: 2010.
-
- Fujikawa K, Chung D, Hendrickson L, Davie E. Amino acid sequence of human factor XI, a blood coagulation factor with four tandem repeats that are highly homologous with plasma prekallikrein. Biochemistry. 1986;25:2417–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL58837/HL/NHLBI NIH HHS/United States
- R01 HL080018/HL/NHLBI NIH HHS/United States
- RG/12/9/29775JE/BHF_/British Heart Foundation/United Kingdom
- R01 HL036365/HL/NHLBI NIH HHS/United States
- R56 HL036365/HL/NHLBI NIH HHS/United States
- HL36365/HL/NHLBI NIH HHS/United States
- R01 HL081326/HL/NHLBI NIH HHS/United States
- RG/07/002/23132/BHF_/British Heart Foundation/United Kingdom
- HL81326/HL/NHLBI NIH HHS/United States
- R01 HL058837/HL/NHLBI NIH HHS/United States
- HL80018/HL/NHLBI NIH HHS/United States
- RG/12/9/29775/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

