Gates of enzymes
- PMID: 23617803
- PMCID: PMC3744840
- DOI: 10.1021/cr300384w
Gates of enzymes
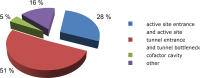
Figures


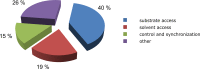
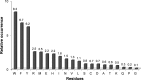
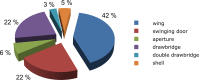
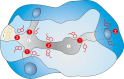
References
-
- Röthlisberger D.; Khersonsky O.; Wollacott A. M.; Jiang L.; DeChancie J.; Betker J.; Gallaher J. L.; Althoff E. A.; Zanghellini A.; Dym O.; Albeck S.; Houk K. N.; Tawfik D. S.; Baker D. Nature 2008, 453, 190. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

