Staphylococcus aureus peptidoglycan stem packing by rotational-echo double resonance NMR spectroscopy
- PMID: 23617832
- PMCID: PMC3796188
- DOI: 10.1021/bi4005039
Staphylococcus aureus peptidoglycan stem packing by rotational-echo double resonance NMR spectroscopy
Abstract
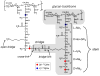
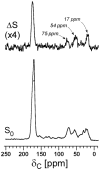
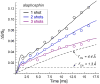
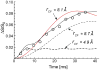
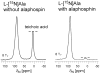
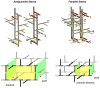
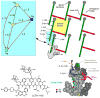
Staphylococcus aureus grown in the presence of an alanine-racemase inhibitor was labeled with d-[1-(13)C]alanine and l-[(15)N]alanine to characterize some details of the peptidoglycan tertiary structure. Rotational-echo double-resonance NMR of intact whole cells was used to measure internuclear distances between (13)C and (15)N of labeled amino acids incorporated in the peptidoglycan, and from those labels to (19)F of a glycopeptide drug specifically bound to the peptidoglycan. The observed (13)C-(15)N average distance of 4.1-4.4 Å between d- and l-alanines in nearest-neighbor peptide stems is consistent with a local, tightly packed, parallel-stem architecture for a repeating structural motif within the peptidoglycan of S. aureus.
Figures
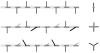

Similar articles
-
Cross-link formation and peptidoglycan lattice assembly in the FemA mutant of Staphylococcus aureus.Biochemistry. 2014 Mar 11;53(9):1420-7. doi: 10.1021/bi4016742. Epub 2014 Feb 26. Biochemistry. 2014. PMID: 24517508 Free PMC article.
-
REDOR constraints on the peptidoglycan lattice architecture of Staphylococcus aureus and its FemA mutant.Biochim Biophys Acta. 2015 Jan;1848(1 Pt B):363-8. doi: 10.1016/j.bbamem.2014.05.025. Epub 2014 Jun 2. Biochim Biophys Acta. 2015. PMID: 24990251 Free PMC article.
-
Conformational and quantitative characterization of oritavancin-peptidoglycan complexes in whole cells of Staphylococcus aureus by in vivo 13C and 15N labeling.J Mol Biol. 2006 Apr 7;357(4):1253-62. doi: 10.1016/j.jmb.2006.01.040. Epub 2006 Jan 30. J Mol Biol. 2006. PMID: 16483598
-
Bacterial cell wall composition and the influence of antibiotics by cell-wall and whole-cell NMR.Philos Trans R Soc Lond B Biol Sci. 2015 Oct 5;370(1679):20150024. doi: 10.1098/rstb.2015.0024. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26370936 Free PMC article. Review.
-
Assembly of the monomer unit of bacterial peptidoglycan.Cell Mol Life Sci. 1998 Apr;54(4):300-4. doi: 10.1007/s000180050155. Cell Mol Life Sci. 1998. PMID: 9614964 Free PMC article. Review.
Cited by
-
Peptidoglycan Compositional Analysis of Enterococcus faecalis Biofilm by Stable Isotope Labeling by Amino Acids in a Bacterial Culture.Biochemistry. 2018 Feb 20;57(7):1274-1283. doi: 10.1021/acs.biochem.7b01207. Epub 2018 Feb 2. Biochemistry. 2018. PMID: 29368511 Free PMC article.
-
Multiparametric AFM reveals turgor-responsive net-like peptidoglycan architecture in live streptococci.Nat Commun. 2015 May 28;6:7193. doi: 10.1038/ncomms8193. Nat Commun. 2015. PMID: 26018339 Free PMC article.
-
Desleucyl-Oritavancin with a Damaged d-Ala-d-Ala Binding Site Inhibits the Transpeptidation Step of Cell-Wall Biosynthesis in Whole Cells of Staphylococcus aureus.Biochemistry. 2017 Mar 14;56(10):1529-1535. doi: 10.1021/acs.biochem.6b01125. Epub 2017 Mar 3. Biochemistry. 2017. PMID: 28221772 Free PMC article.
-
Dual Mode of Action for Plusbacin A3 in Staphylococcus aureus.J Phys Chem B. 2017 Feb 23;121(7):1499-1505. doi: 10.1021/acs.jpcb.6b11039. Epub 2017 Feb 13. J Phys Chem B. 2017. PMID: 28135800 Free PMC article.
-
Solid-state NMR Reveals the Carbon-based Molecular Architecture of Cryptococcus neoformans Fungal Eumelanins in the Cell Wall.J Biol Chem. 2015 May 29;290(22):13779-90. doi: 10.1074/jbc.M114.618389. Epub 2015 Mar 30. J Biol Chem. 2015. PMID: 25825492 Free PMC article.
References
-
- Rogers HJ, Ward JB, Perkins HR. Microbial cell walls and membranes. Chapman and Hall; London; New York: 1980.
-
- van Heijenoort J. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology. 2001;11:25R–36R. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

