DNA binding specificities of the long zinc-finger recombination protein PRDM9
- PMID: 23618393
- PMCID: PMC4053984
- DOI: 10.1186/gb-2013-14-4-r35
DNA binding specificities of the long zinc-finger recombination protein PRDM9
Abstract
Background: Meiotic recombination ensures proper segregation of homologous chromosomes and creates genetic variation. In many organisms, recombination occurs at limited sites, termed 'hotspots', whose positions in mammals are determined by PR domain member 9 (PRDM9), a long-array zinc-finger and chromatin-modifier protein. Determining the rules governing the DNA binding of PRDM9 is a major issue in understanding how it functions.
Results: Mouse PRDM9 protein variants bind to hotspot DNA sequences in a manner that is specific for both PRDM9 and DNA haplotypes, and that in vitro binding parallels its in vivo biological activity. Examining four hotspots, three activated by Prdm9Cst and one activated by Prdm9Dom2, we found that all binding sites required the full array of 11 or 12 contiguous fingers, depending on the allele, and that there was little sequence similarity between the binding sites of the three Prdm9Cst activated hotspots. The binding specificity of each position in the Hlx1 binding site, activated by Prdm9Cst, was tested by mutating each nucleotide to its three alternatives. The 31 positions along the binding site varied considerably in the ability of alternative bases to support binding, which also implicates a role for additional binding to the DNA phosphate backbone.
Conclusions: These results, which provide the first detailed mapping of PRDM9 binding to DNA and, to our knowledge, the most detailed analysis yet of DNA binding by a long zinc-finger array, make clear that the binding specificities of PRDM9, and possibly other long-array zinc-finger proteins, are unusually complex.
Figures


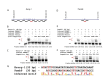
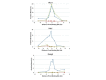
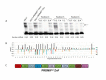
Comment in
-
The complex binding of PRDM9.Genome Biol. 2013 Apr 24;14(4):112. doi: 10.1186/gb-2013-14-4-112. Genome Biol. 2013. PMID: 23651476 Free PMC article.
References
-
- Drouaud J, Camilleri C, Bourguignon PY, Canaguier A, Berard A, Vezon D, Giancola S, Brunel D, Colot V, Prum B, Quesneville H, Mézard C. Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination "hot spots". Genome Res. 2006;14:106–114. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

