Role of MerH in mercury resistance in the archaeon Sulfolobus solfataricus
- PMID: 23619003
- PMCID: PMC3709690
- DOI: 10.1099/mic.0.065854-0
Role of MerH in mercury resistance in the archaeon Sulfolobus solfataricus
Abstract
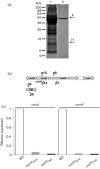
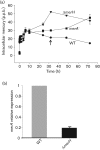
Crenarchaeota include extremely thermoacidophilic organisms that thrive in geothermal environments dominated by sulfidic ores and heavy metals such as mercury. Mercuric ion, Hg(II), inactivates transcription in the crenarchaeote Sulfolobus solfataricus and simultaneously derepresses transcription of a resistance operon, merHAI, through interaction with the MerR transcription factor. While mercuric reductase (MerA) is required for metal resistance, the role of MerH, an adjacent small and predicted product of an ORF, has not been explored. Inactivation of MerH either by nonsense mutation or by in-frame deletion diminished Hg(II) resistance of mutant cells. Promoter mapping studies indicated that Hg(II) sensitivity of the merH nonsense mutant arose through transcriptional polarity, and its metal resistance was restored partially by single copy merH complementation. Since MerH was not required in vitro for MerA-catalysed Hg(II) reduction, MerH may play an alternative role in metal resistance. Inductively coupled plasma-mass spectrometry analysis of the MerH deletion strain following metal challenge indicated that there was prolonged retention of intracellular Hg(II). Finally, a reduced rate of mer operon induction in the merH deletion mutant suggested that the requirement for MerH could result from metal trafficking to the MerR transcription factor.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

