MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment
- PMID: 23620378
- PMCID: PMC3749523
- DOI: 10.1099/vir.0.052910-0
MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment
Abstract
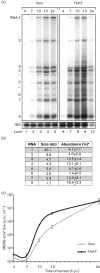
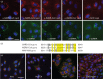
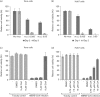
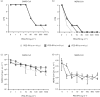
Coronavirus (CoV) infections are commonly associated with respiratory and enteric disease in humans and animals. The 2003 outbreak of severe acute respiratory syndrome (SARS) highlighted the potentially lethal consequences of CoV-induced disease in humans. In 2012, a novel CoV (Middle East Respiratory Syndrome coronavirus; MERS-CoV) emerged, causing 49 human cases thus far, of which 23 had a fatal outcome. In this study, we characterized MERS-CoV replication and cytotoxicity in human and monkey cell lines. Electron microscopy of infected Vero cells revealed extensive membrane rearrangements, including the formation of double-membrane vesicles and convoluted membranes, which have been implicated previously in the RNA synthesis of SARS-CoV and other CoVs. Following infection, we observed rapidly increasing viral RNA synthesis and release of high titres of infectious progeny, followed by a pronounced cytopathology. These characteristics were used to develop an assay for antiviral compound screening in 96-well format, which was used to identify cyclosporin A as an inhibitor of MERS-CoV replication in cell culture. Furthermore, MERS-CoV was found to be 50-100 times more sensitive to alpha interferon (IFN-α) treatment than SARS-CoV, an observation that may have important implications for the treatment of MERS-CoV-infected patients. MERS-CoV infection did not prevent the IFN-induced nuclear translocation of phosphorylated STAT1, in contrast to infection with SARS-CoV where this block inhibits the expression of antiviral genes. These findings highlight relevant differences between these distantly related zoonotic CoVs in terms of their interaction with and evasion of the cellular innate immune response.
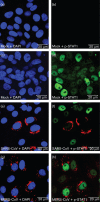
Figures


References
-
- Brockway S. M., Clay C. T., Lu X. T., Denison M. R. (2003). Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J Virol 77, 10515–10527 10.1128/JVI.77.19.10515-10527.2003 - DOI - PMC - PubMed
-
- Cameron M. J., Kelvin A. A., Leon A. J., Cameron C. M., Ran L., Xu L., Chu Y. K., Danesh A., Fang Y. & other authors (2012). Lack of innate interferon responses during SARS coronavirus infection in a vaccination and reinfection ferret model. PLoS ONE 7, e45842 10.1371/journal.pone.0045842 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

