Nonsense-mediated mRNA decay controls the changes in yeast ribosomal protein pre-mRNAs levels upon osmotic stress
- PMID: 23620734
- PMCID: PMC3631235
- DOI: 10.1371/journal.pone.0061240
Nonsense-mediated mRNA decay controls the changes in yeast ribosomal protein pre-mRNAs levels upon osmotic stress
Abstract
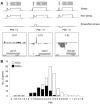
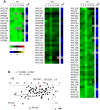
The expression of ribosomal protein (RP) genes requires a substantial part of cellular transcription, processing and translation resources. Thus, the RP expression must be tightly regulated in response to conditions that compromise cell survival. In Saccharomyces cerevisiae cells, regulation of the RP gene expression at the transcriptional, mature mRNA stability and translational levels during the response to osmotic stress has been reported. Reprogramming global protein synthesis upon osmotic shock includes the movement of ribosomes from RP transcripts to stress-induced mRNAs. Using tiling arrays, we show that osmotic stress yields a drop in the levels of RP pre-mRNAs in S. cerevisiae cells. An analysis of the tiling array data, together with transcription rates data, shows a poor correlation, indicating that the drop in the RP pre-mRNA levels is not merely a result of the lowered RP transcription rates. A kinetic study using quantitative RT-PCR confirmed the decrease in the levels of several RP-unspliced transcripts during the first 15 minutes of osmotic stress, which seems independent of MAP kinase Hog1. Moreover, we found that the mutations in the components of the nonsense-mediated mRNA decay (NMD), Upf1, Upf2, Upf3 or in exonuclease Xrn1, eliminate the osmotic stress-induced drop in RP pre-mRNAs. Altogether, our results indicate that the degradation of yeast RP unspliced transcripts by NMD increases during osmotic stress, and suggest that this might be another mechanism to control RP synthesis during the stress response.
Conflict of interest statement
Figures



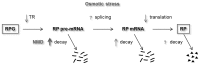
Similar articles
-
Sequential RNA degradation pathways provide a fail-safe mechanism to limit the accumulation of unspliced transcripts in Saccharomyces cerevisiae.RNA. 2012 Aug;18(8):1563-72. doi: 10.1261/rna.033779.112. Epub 2012 Jul 2. RNA. 2012. PMID: 22753783 Free PMC article.
-
Widespread impact of nonsense-mediated mRNA decay on the yeast intronome.Mol Cell. 2008 Aug 8;31(3):360-70. doi: 10.1016/j.molcel.2008.07.005. Mol Cell. 2008. PMID: 18691968 Free PMC article.
-
Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast.Proc Natl Acad Sci U S A. 2005 Mar 22;102(12):4258-63. doi: 10.1073/pnas.0500684102. Epub 2005 Mar 7. Proc Natl Acad Sci U S A. 2005. PMID: 15753296 Free PMC article.
-
NMD monitors translational fidelity 24/7.Curr Genet. 2017 Dec;63(6):1007-1010. doi: 10.1007/s00294-017-0709-4. Epub 2017 May 23. Curr Genet. 2017. PMID: 28536849 Free PMC article. Review.
-
Regulation of natural mRNAs by the nonsense-mediated mRNA decay pathway.Eukaryot Cell. 2014 Sep;13(9):1126-35. doi: 10.1128/EC.00090-14. Epub 2014 Jul 18. Eukaryot Cell. 2014. PMID: 25038084 Free PMC article. Review.
Cited by
-
The Lsm1-7/Pat1 complex binds to stress-activated mRNAs and modulates the response to hyperosmotic shock.PLoS Genet. 2018 Jul 30;14(7):e1007563. doi: 10.1371/journal.pgen.1007563. eCollection 2018 Jul. PLoS Genet. 2018. PMID: 30059503 Free PMC article.
-
Three Microbial Musketeers of the Seas: Shewanella baltica, Aliivibrio fischeri and Vibrio harveyi, and Their Adaptation to Different Salinity Probed by a Proteomic Approach.Int J Mol Sci. 2022 Jan 6;23(2):619. doi: 10.3390/ijms23020619. Int J Mol Sci. 2022. PMID: 35054801 Free PMC article.
-
Yeast Translation Elongation Factor eIF5A Expression Is Regulated by Nutrient Availability through Different Signalling Pathways.Int J Mol Sci. 2020 Dec 28;22(1):219. doi: 10.3390/ijms22010219. Int J Mol Sci. 2020. PMID: 33379337 Free PMC article.
-
Fertility and polarized cell growth depends on eIF5A for translation of polyproline-rich formins in Saccharomyces cerevisiae.Genetics. 2014 Aug;197(4):1191-200. doi: 10.1534/genetics.114.166926. Epub 2014 Jun 11. Genetics. 2014. PMID: 24923804 Free PMC article.
-
Evolutionary conserved role of eukaryotic translation factor eIF5A in the regulation of actin-nucleating formins.Sci Rep. 2017 Aug 29;7(1):9580. doi: 10.1038/s41598-017-10057-y. Sci Rep. 2017. PMID: 28852021 Free PMC article.
References
-
- Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends in biochemical sciences 24: 437–440. - PubMed
-
- Parenteau J, Durand M, Morin G, Gagnon J, Lucier JF, et al. (2011) Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell 147: 320–331. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

