Structural and functional insights into Saccharomyces cerevisiae riboflavin biosynthesis reductase RIB7
- PMID: 23620735
- PMCID: PMC3631187
- DOI: 10.1371/journal.pone.0061249
Structural and functional insights into Saccharomyces cerevisiae riboflavin biosynthesis reductase RIB7
Abstract
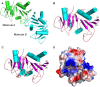
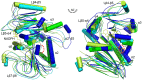
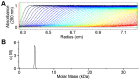
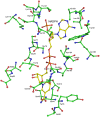
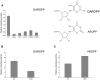
Saccharomyces cerevisiae RIB7 (ScRIB7) is a potent target for anti-fungal agents because of its involvement in the riboflavin biosynthesis pathway as a NADPH-dependent reductase. However, the catalytic mechanism of riboflavin biosynthesis reductase (RBSRs) is controversial, and enzyme structure information is still lacking in eukaryotes. Here we report the crystal structure of Saccharomyces cerevisiae RIB7 at 2.10 Å resolution and its complex with NADPH at 2.35 Å resolution. ScRIB7 exists as a stable homodimer, and each subunit consists of nine central β-sheets flanked by five helices, resembling the structure of RIB7 homologues. A conserved G(76)-X-G(78)-Xn-G(181)-G(182) motif is present at the NADPH pyrophosphate group binding site. Activity assays confirmed the necessity of Thr79, Asp83, Glu180 and Gly182 for the activity of ScRIB7. Substrate preference of ScRIB7 was altered by mutating one residue (Thr35) to a Lysine, implying that ScRIB7 Thr35 and its corresponding residue, a lysine in bacteria, are important in substrate-specific recognition.
Conflict of interest statement
Figures

Similar articles
-
Evolution of archaeal Rib7 and eubacterial RibG reductases in riboflavin biosynthesis: Substrate specificity and cofactor preference.Biochem Biophys Res Commun. 2018 Sep 3;503(1):195-201. doi: 10.1016/j.bbrc.2018.06.002. Epub 2018 Jun 8. Biochem Biophys Res Commun. 2018. PMID: 29864427
-
Structural insights into the cofactor-assisted substrate recognition of yeast methylglyoxal/isovaleraldehyde reductase Gre2.Biochim Biophys Acta. 2014 Sep;1844(9):1486-92. doi: 10.1016/j.bbapap.2014.05.008. Epub 2014 May 27. Biochim Biophys Acta. 2014. PMID: 24879127
-
Structures of Saccharomyces cerevisiae D-arabinose dehydrogenase Ara1 and its complex with NADPH: implications for cofactor-assisted substrate recognition.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 Nov;69(Pt 11):1190-5. doi: 10.1107/S1744309113026857. Epub 2013 Oct 26. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 24192347 Free PMC article.
-
Structural insights into the cofactor-assisted substrate recognition of yeast quinone oxidoreductase Zta1.J Struct Biol. 2011 Oct;176(1):112-8. doi: 10.1016/j.jsb.2011.07.010. Epub 2011 Jul 26. J Struct Biol. 2011. PMID: 21820057
-
Direct enzyme assay evidence confirms aldehyde reductase function of Ydr541cp and Ygl039wp from Saccharomyces cerevisiae.Yeast. 2015 Apr;32(4):399-407. doi: 10.1002/yea.3067. Epub 2015 Feb 26. Yeast. 2015. PMID: 25656103
Cited by
-
The Riboflavin Metabolism in Four Saccharomyces cerevisiae Wine Strains: Assessment in Oenological Condition and Potential Implications with the Light-Struck Taste.J Fungi (Basel). 2023 Jan 5;9(1):78. doi: 10.3390/jof9010078. J Fungi (Basel). 2023. PMID: 36675899 Free PMC article.
-
Structural Insights into Mycobacterium tuberculosis Rv2671 Protein as a Dihydrofolate Reductase Functional Analogue Contributing to para-Aminosalicylic Acid Resistance.Biochemistry. 2016 Feb 23;55(7):1107-19. doi: 10.1021/acs.biochem.5b00993. Epub 2016 Feb 5. Biochemistry. 2016. PMID: 26848874 Free PMC article.
References
-
- Macheroux P, Kappes B, Ealick SE (2011) Flavogenomics–a genomic and structural view of flavin-dependent proteins. FEBS J 278: 2625–2634. - PubMed
-
- Takami Y, Gong H, Amemiya T (2004) Riboflavin deficiency induces ocular surface damage. Ophthalmic Res 36: 156–165. - PubMed
-
- Hedtke B, Alawady A, Albacete A, Kobayashi K, Melzer M, et al. (2012) Deficiency in riboflavin biosynthesis affects tetrapyrrole biosynthesis in etiolated Arabidopsis tissue. Plant Mol Biol 78: 77–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

