The Drosophila BTB domain protein Jim Lovell has roles in multiple larval and adult behaviors
- PMID: 23620738
- PMCID: PMC3631165
- DOI: 10.1371/journal.pone.0061270
The Drosophila BTB domain protein Jim Lovell has roles in multiple larval and adult behaviors
Erratum in
- PLoS One. 2013;8(8). doi:10.1371/annotation/88d89372-9fdc-48d7-83b3-26b61142a5e2
Abstract
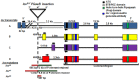
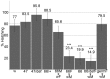
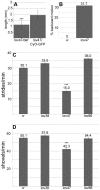
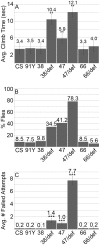
Innate behaviors have their origins in the specification of neural fates during development. Within Drosophila, BTB (Bric-a-brac,Tramtrack, Broad) domain proteins such as Fruitless are known to play key roles in the neural differentiation underlying such responses. We previously identified a gene, which we have termed jim lovell (lov), encoding a BTB protein with a role in gravity responses. To understand more fully the behavioral roles of this gene we have investigated its function through several approaches. Transcript and protein expression patterns have been examined and behavioral phenotypes of new lov mutations have been characterized. Lov is a nuclear protein, suggesting a role as a transcriptional regulator, as for other BTB proteins. In late embryogenesis, Lov is expressed in many CNS and PNS neurons. An examination of the PNS expression indicates that lov functions in the late specification of several classes of sensory neurons. In particular, only two of the five abdominal lateral chordotonal neurons express Lov, predicting functional variation within this highly similar group. Surprisingly, Lov is also expressed very early in embryogenesis in ways that suggests roles in morphogenetic movements, amnioserosa function and head neurogenesis. The phenotypes of two new lov mutations that delete adjacent non-coding DNA regions are strikingly different suggesting removal of different regulatory elements. In lov(47) , Lov expression is lost in many embryonic neurons including the two lateral chordotonal neurons. lov(47) mutant larvae show feeding and locomotor defects including spontaneous backward movement. Adult lov(47) males perform aberrant courtship behavior distinguished by courtship displays that are not directed at the female. lov(47) adults also show more defective negative gravitaxis than the previously isolated lov(91Y) mutant. In contrast, lov(66) produces largely normal behavior but severe female sterility associated with ectopic lov expression in the ovary. We propose a negative regulatory role for the DNA deleted in lov(66) .
Conflict of interest statement
Figures





Similar articles
-
Failure to Burrow and Tunnel Reveals Roles for jim lovell in the Growth and Endoreplication of the Drosophila Larval Tracheae.PLoS One. 2016 Aug 5;11(8):e0160233. doi: 10.1371/journal.pone.0160233. eCollection 2016. PLoS One. 2016. PMID: 27494251 Free PMC article.
-
The roles of jim lovell and uninflatable in different endopolyploid larval tissues of Drosophila melanogaster.PLoS One. 2020 Aug 21;15(8):e0237662. doi: 10.1371/journal.pone.0237662. eCollection 2020. PLoS One. 2020. PMID: 32822370 Free PMC article.
-
The Drosophila CPEB protein Orb2 has a novel expression pattern and is important for asymmetric cell division and nervous system function.Genetics. 2011 Nov;189(3):907-21. doi: 10.1534/genetics.110.123646. Epub 2011 Sep 6. Genetics. 2011. PMID: 21900268 Free PMC article.
-
What does the fruitless gene tell us about nature vs. nurture in the sex life of Drosophila?Fly (Austin). 2017 Apr 3;11(2):139-147. doi: 10.1080/19336934.2016.1263778. Epub 2016 Nov 23. Fly (Austin). 2017. PMID: 27880074 Free PMC article. Review.
-
Neurogenetics of courtship and mating in Drosophila.Adv Genet. 2008;62:67-184. doi: 10.1016/S0065-2660(08)00603-2. Adv Genet. 2008. PMID: 19010254 Review.
Cited by
-
Expression Analysis Reveals the Association of Several Genes with Pupal Diapause in Bactrocera minax (Diptera: Tephritidae).Insects. 2019 Jun 13;10(6):169. doi: 10.3390/insects10060169. Insects. 2019. PMID: 31200584 Free PMC article.
-
Dissecting the enhancer gene regulatory network in early Drosophila spermatogenesis.Nat Commun. 2025 Jul 23;16(1):6766. doi: 10.1038/s41467-025-62046-9. Nat Commun. 2025. PMID: 40695849 Free PMC article.
-
Patterns of Population Structure and Introgression Among Recently Differentiated Drosophila melanogaster Populations.Mol Biol Evol. 2022 Nov 3;39(11):msac223. doi: 10.1093/molbev/msac223. Mol Biol Evol. 2022. PMID: 36251862 Free PMC article.
-
Reduction in Gonad Development and Sperm Motility in Male Brown Planthopper Nilaparvata lugens via RNAi-Mediated Knockdown of tramtrack.Int J Mol Sci. 2025 Apr 12;26(8):3643. doi: 10.3390/ijms26083643. Int J Mol Sci. 2025. PMID: 40332247 Free PMC article.
-
Selection and Validation of Reference Genes for RT-qPCR Analysis of the Ladybird Beetle Henosepilachna vigintioctomaculata.Front Physiol. 2018 Nov 14;9:1614. doi: 10.3389/fphys.2018.01614. eCollection 2018. Front Physiol. 2018. PMID: 30483159 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

