Spatio-temporal distribution of Smads and role of Smads/TGF-β/BMP-4 in the regulation of mouse bladder organogenesis
- PMID: 23620745
- PMCID: PMC3631207
- DOI: 10.1371/journal.pone.0061340
Spatio-temporal distribution of Smads and role of Smads/TGF-β/BMP-4 in the regulation of mouse bladder organogenesis
Abstract
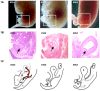
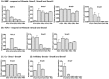
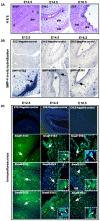
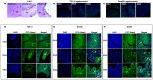
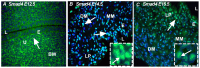
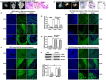
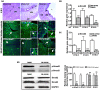
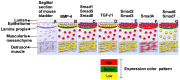
Although Shh, TGF-β and BMP-4 regulate radial patterning of the bladder mesenchyme and smooth muscle differentiation, it is not known what transcription factors, local environmental cues or signaling cascades mediate bladder smooth muscle differentiation. We investigated the expression patterns of signaling mediated by Smad2 and Smad3 in the mouse embryonic bladder from E12.5 to E16.5 by using qRT-PCR, in situ hybridization and antibodies specifically recognizing individual Smad proteins. The role of Smad2 and Smad3 during smooth muscle formation was examined by disrupting the Smad2/3 signaling pathway using TβR1 inhibitor SB-431542 in organ culture system. qRT-PCR results showed that R-Smads, Co-Smad and I-Smads were all expressed during bladder development. RNA ISH for BMP-4 and immunostaining of TGF-β1 showed that BMP-4 and TGF-β1 were expressed in the transitional epithelium, lamina propia and muscularis mucosa. Smad1, Smad5 and Smad8 were first expressed in the bladder epithelium and continued to be expressed in the transitional epithelium, muscularis mesenchyme and lamina propia as the bladder developed. Smad2, Smad3 and Smad4 were first detected in the bladder epithelium and subsequently were expressed in the muscularis mesenchyme and lamina propia. Smad6 and Smad7 showed overlapping expression with R-Smads, which are critical for bladder development. In bladder explants (E12.5 to E16.5) culture, Smad2 and Smad3 were found localized within the nuclei, suggesting critical transcriptional regulatory effects during bladder development. E12.5 to E16.5 bladders were cultured with and without TβR1 inhibitor SB-431542 and assessed by qRT-PCR and immunofluorescence. After three days in culture in SB-431542, α-SMA, Smad2 and Smad3 expressions were significantly decreased compared with controls, however, with no significant changes in the expression of smooth muscle myosin heavy chain (SM-Myh. Based on the Smad expression patterns, we suggest that individual or combinations of Smads may be necessary during mouse bladder organogenesis and may be critical mediators for bladder smooth muscle differentiation.
Conflict of interest statement
Figures


Similar articles
-
Signalling molecules involved in mouse bladder smooth muscle cellular differentiation.Int J Dev Biol. 2010;54(1):175-80. doi: 10.1387/ijdb.082610bl. Int J Dev Biol. 2010. PMID: 20013655 Free PMC article.
-
Transforming growth factor-β(1) represses bone morphogenetic protein-mediated Smad signaling in pulmonary artery smooth muscle cells via Smad3.Am J Respir Cell Mol Biol. 2013 Dec;49(6):1135-45. doi: 10.1165/rcmb.2012-0470OC. Am J Respir Cell Mol Biol. 2013. PMID: 23937428 Free PMC article.
-
Developmental expression of Smad1-7 suggests critical function of TGF-beta/BMP signaling in regulating epithelial-mesenchymal interaction during tooth morphogenesis.Int J Dev Biol. 2003 Feb;47(1):31-9. Int J Dev Biol. 2003. PMID: 12653249
-
[The role of Smads and related transcription factors in the signal transduction of bone morphogenetic protein inducing bone formation].Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2003 Sep;17(5):359-62. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2003. PMID: 14551929 Review. Chinese.
-
Diverse roles of TGF-β/Smads in renal fibrosis and inflammation.Int J Biol Sci. 2011;7(7):1056-67. doi: 10.7150/ijbs.7.1056. Epub 2011 Sep 2. Int J Biol Sci. 2011. PMID: 21927575 Free PMC article. Review.
Cited by
-
Modulation of phosphatase of regenerating liver-1 within placental mesenchymal stem cells instigates the transition between epithelial-to-mesenchymal transition and mesenchymal-to-epithelial transition subsequent to hepatic fibrosis.Clin Mol Hepatol. 2025 Jul;31(3):823-840. doi: 10.3350/cmh.2024.0741. Epub 2025 Jan 22. Clin Mol Hepatol. 2025. PMID: 39838827 Free PMC article.
-
Migration pathways of sacral neural crest during development of lower urogenital tract innervation.Dev Biol. 2017 Sep 1;429(1):356-369. doi: 10.1016/j.ydbio.2017.04.011. Epub 2017 Apr 25. Dev Biol. 2017. PMID: 28449850 Free PMC article.
-
The Interactivity between TGFβ and BMP Signaling in Organogenesis, Fibrosis, and Cancer.Cells. 2019 Sep 23;8(10):1130. doi: 10.3390/cells8101130. Cells. 2019. PMID: 31547567 Free PMC article. Review.
-
Fgfr2 is integral for bladder mesenchyme patterning and function.Am J Physiol Renal Physiol. 2017 Apr 1;312(4):F607-F618. doi: 10.1152/ajprenal.00463.2016. Epub 2017 Jan 4. Am J Physiol Renal Physiol. 2017. PMID: 28052872 Free PMC article.
-
TGF-β1 induces EMT reprogramming of porcine bladder urothelial cells into collagen producing fibroblasts-like cells in a Smad2/Smad3-dependent manner.J Cell Commun Signal. 2014 Mar;8(1):39-58. doi: 10.1007/s12079-013-0216-4. Epub 2013 Dec 12. J Cell Commun Signal. 2014. PMID: 24338442 Free PMC article.
References
-
- Li J, Shiroyanagi Y, Lin G, Haqq C, Lin CS, et al. (2006) Serum response factor, its cofactors, and epithelial-mesenchymal signaling in urinary bladder smooth muscle formation. Differentiation 74: 30–39. - PubMed
-
- Shiroyanagi Y, Liu B, Cao M, Agras K, Li J, et al. (2007) Urothelial sonic hedgehog signaling plays an important role in bladder smooth muscle formation. Differentiation 75: 968–977. - PubMed
-
- Baskin LS, Hayward SW, Young P, Cunha GR (1996) Role of mesenchymal-epithelial interaction in normal bladder development. J Urol 156: 1820–1827. - PubMed
-
- Baskin LS, Sutherland RA, DiSandro MS, Thompson AA, Cunha GR (1997) Cellular signaling in the bladder. Frontiers in Bioscience 2: 592–595. - PubMed
-
- Baskin LS, Hayward SW, Young P, Cunha GR (1996a) Role of mesenchymal-epithelial interaction in bladder development. J Urol 156: 1820–1827. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

