Chlamydia trachomatis infection induces replication of latent HHV-6
- PMID: 23620749
- PMCID: PMC3631192
- DOI: 10.1371/journal.pone.0061400
Chlamydia trachomatis infection induces replication of latent HHV-6
Abstract
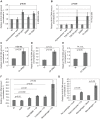
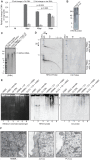
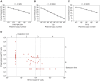
Human herpesvirus-6 (HHV-6) exists in latent form either as a nuclear episome or integrated into human chromosomes in more than 90% of healthy individuals without causing clinical symptoms. Immunosuppression and stress conditions can reactivate HHV-6 replication, associated with clinical complications and even death. We have previously shown that co-infection of Chlamydia trachomatis and HHV-6 promotes chlamydial persistence and increases viral uptake in an in vitro cell culture model. Here we investigated C. trachomatis-induced HHV-6 activation in cell lines and fresh blood samples from patients having Chromosomally integrated HHV-6 (CiHHV-6). We observed activation of latent HHV-6 DNA replication in CiHHV-6 cell lines and fresh blood cells without formation of viral particles. Interestingly, we detected HHV-6 DNA in blood as well as cervical swabs from C. trachomatis-infected women. Low virus titers correlated with high C. trachomatis load and vice versa, demonstrating a potentially significant interaction of these pathogens in blood cells and in the cervix of infected patients. Our data suggest a thus far underestimated interference of HHV-6 and C. trachomatis with a likely impact on the disease outcome as consequence of co-infection.
Conflict of interest statement
Figures
Similar articles
-
Reactivation of chromosomally integrated human herpesvirus-6 by telomeric circle formation.PLoS Genet. 2013;9(12):e1004033. doi: 10.1371/journal.pgen.1004033. Epub 2013 Dec 19. PLoS Genet. 2013. PMID: 24367281 Free PMC article.
-
The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro.Proc Natl Acad Sci U S A. 2010 Mar 23;107(12):5563-8. doi: 10.1073/pnas.0913586107. Epub 2010 Mar 8. Proc Natl Acad Sci U S A. 2010. PMID: 20212114 Free PMC article.
-
Chromosomally integrated human herpesvirus 6 in heart failure: prevalence and treatment.Eur J Heart Fail. 2015 Jan;17(1):9-19. doi: 10.1002/ejhf.194. Epub 2014 Nov 11. Eur J Heart Fail. 2015. PMID: 25388833
-
Chromosomally integrated human herpesvirus 6: questions and answers.Rev Med Virol. 2012 May;22(3):144-55. doi: 10.1002/rmv.715. Epub 2011 Nov 4. Rev Med Virol. 2012. PMID: 22052666 Free PMC article. Review.
-
Chromosomally Integrated Human Herpesvirus 6: Models of Viral Genome Release from the Telomere and Impacts on Human Health.Viruses. 2017 Jul 12;9(7):184. doi: 10.3390/v9070184. Viruses. 2017. PMID: 28704957 Free PMC article. Review.
Cited by
-
A "plus one" strategy impacts replication of felid alphaherpesvirus 1, Mycoplasma and Chlamydia, and the metabolism of coinfected feline cells.mSystems. 2024 Oct 22;9(10):e0085224. doi: 10.1128/msystems.00852-24. Epub 2024 Sep 24. mSystems. 2024. PMID: 39315777 Free PMC article.
-
When Bacteria and Viruses Collide: A Tale of Chlamydia trachomatis and Sexually Transmitted Viruses.Viruses. 2023 Sep 19;15(9):1954. doi: 10.3390/v15091954. Viruses. 2023. PMID: 37766360 Free PMC article. Review.
-
Reactivation of chromosomally integrated human herpesvirus-6 by telomeric circle formation.PLoS Genet. 2013;9(12):e1004033. doi: 10.1371/journal.pgen.1004033. Epub 2013 Dec 19. PLoS Genet. 2013. PMID: 24367281 Free PMC article.
-
Roseolovirus molecular biology: recent advances.Curr Opin Virol. 2014 Dec;9:170-7. doi: 10.1016/j.coviro.2014.10.004. Epub 2014 Nov 27. Curr Opin Virol. 2014. PMID: 25437229 Free PMC article. Review.
-
Detection of Chlamydiaceae and Chlamydia-like organisms on the ocular surface of children and adults from a trachoma-endemic region.Sci Rep. 2018 May 9;8(1):7432. doi: 10.1038/s41598-018-23887-1. Sci Rep. 2018. PMID: 29743637 Free PMC article.
References
-
- Lindquester GJ, Pellett PE (1991) Properties of the human herpesvirus 6 strain Z29 genome: G+C content, length, and presence of variable-length directly repeated terminal sequence elements. Virol 182: 102–110. - PubMed
-
- Tanaka-Taya K, Sashihara J, Kurahashi H, Amo K, Miyagawa H, et al. (2004) Human herpesvirus 6 (HHV-6) is transmitted from parent to child in an integrated form and characterization of cases with chromosomally integrated HHV-6 DNA. J Med Virol 73: 465–473. - PubMed
-
- Daibata M, Taguchi T, Nemoto Y, Taguchi H, Miyoshi I (1999) Inheritance of chromosomally integrated human herpesvirus 6 DNA. Blood 94: 1545. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

