Indoor airborne bacterial communities are influenced by ventilation, occupancy, and outdoor air source
- PMID: 23621155
- PMCID: PMC4285785
- DOI: 10.1111/ina.12047
Indoor airborne bacterial communities are influenced by ventilation, occupancy, and outdoor air source
Abstract
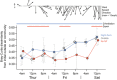
Architects and engineers are beginning to consider a new dimension of indoor air: the structure and composition of airborne microbial communities. A first step in this emerging field is to understand the forces that shape the diversity of bioaerosols across space and time within the built environment. In an effort to elucidate the relative influences of three likely drivers of indoor bioaerosol diversity - variation in outdoor bioaerosols, ventilation strategy, and occupancy load - we conducted an intensive temporal study of indoor airborne bacterial communities in a high-traffic university building with a hybrid HVAC (mechanically and naturally ventilated) system. Indoor air communities closely tracked outdoor air communities, but human-associated bacterial genera were more than twice as abundant in indoor air compared with outdoor air. Ventilation had a demonstrated effect on indoor airborne bacterial community composition; changes in outdoor air communities were detected inside following a time lag associated with differing ventilation strategies relevant to modern building design. Our results indicate that both occupancy patterns and ventilation strategies are important for understanding airborne microbial community dynamics in the built environment.
Keywords: Airborne bacterial community; Bioaerosol; Built environment; Indoor microbial ecology; Natural ventilation.
© 2013 The Authors. Indoor Air published by John Wiley & Sons Ltd.
Figures



References
-
- Bowers RM, McCubbin IB, Hallar AG, Fierer N. Seasonal variability in airborne bacterial communities at a high-elevation site. Atmos. Environ. 2012;50:41–49.
-
- Brown GZ, DeKay M. Sun, Wind & Light: Architectural Design Strategies. 2nd edn. New York, NY: John Wiley & Sons, Inc; 2001.
-
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

