Silencing of Nicotiana benthamiana Neuroblastoma-Amplified Gene causes ER stress and cell death
- PMID: 23621803
- PMCID: PMC3654999
- DOI: 10.1186/1471-2229-13-69
Silencing of Nicotiana benthamiana Neuroblastoma-Amplified Gene causes ER stress and cell death
Abstract
Background: Neuroblastoma Amplified Gene (NAG) was identified as a gene co-amplified with the N-myc gene, whose genomic amplification correlates with poor prognosis of neuroblastoma. Later it was found that NAG is localized in endoplasmic reticulum (ER) and is a component of the syntaxin 18 complex that is involved in Golgi-to-ER retrograde transport in human cells. Homologous sequences of NAG are found in plant databases, but its function in plant cells remains unknown.
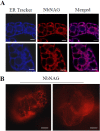
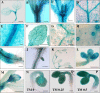
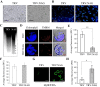
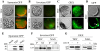
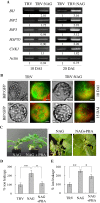
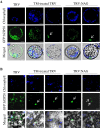
Results: Nicotiana benthamania Neuroblastoma-Amplified Gene (NbNAG) encodes a protein of 2,409 amino acids that contains the secretory pathway Sec39 domain and is mainly localized in the ER. Silencing of NbNAG by virus-induced gene silencing resulted in growth arrest and acute plant death with morphological markers of programmed cell death (PCD), which include chromatin fragmentation and modification of mitochondrial membrane potential. NbNAG deficiency caused induction of ER stress genes, disruption of the ER network, and relocation of bZIP28 transcription factor from the ER membrane to the nucleus, similar to the phenotypes of tunicamycin-induced ER stress in a plant cell. NbNAG silencing caused defects in intracellular transport of diverse cargo proteins, suggesting that a blocked secretion pathway by NbNAG deficiency causes ER stress and programmed cell death.
Conclusions: These results suggest that NAG, a conserved protein from yeast to mammals, plays an essential role in plant growth and development by modulating protein transport pathway, ER stress response and PCD.
Figures

References
-
- Wertz IE, Hanley MR. Diverse molecular provocation of programmed cell death. Trends Biochem Sci. 1996;21:359–364. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

