Incorporating phylogenetic-based covarying mutations into RNAalifold for RNA consensus structure prediction
- PMID: 23621982
- PMCID: PMC3691524
- DOI: 10.1186/1471-2105-14-142
Incorporating phylogenetic-based covarying mutations into RNAalifold for RNA consensus structure prediction
Abstract
Background: RNAalifold, a popular computational method for RNA consensus structure prediction, incorporates covarying mutations into a thermodynamic model to fold the aligned RNA sequences. When quantifying covariance, it evaluates conserved signals of two aligned columns with base-pairing rules. This scoring scheme performs better than some other approaches, such as mutual information. However it ignores the phylogenetic history of the aligned sequences, which is an important criterion to evaluate the level of sequence covariance.
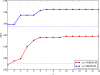
Results: In this article, in order to improve the accuracy of consensus structure folding, we propose a novel approach named PhyloRNAalifold. It incorporates the number of covarying mutations on the phylogenetic tree of the aligned sequences into the covariance scoring of RNAalifold. The benchmarking results show that the new scoring scheme of PhyloRNAalifold can improve the consensus structure detection of RNAalifold.
Conclusion: Incorporating additional phylogenetic information of aligned sequences into the covariance scoring of RNAalifold can improve its performance of consensus structures folding. This improvement is correlated with alignment characteristics, such as pair-wise identity and the number of sequences in the alignment.
Figures

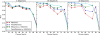
Similar articles
-
Phylogenetic and Chemical Probing Information as Soft Constraints in RNA Secondary Structure Prediction.J Comput Biol. 2024 Jun;31(6):549-563. doi: 10.1089/cmb.2024.0519. J Comput Biol. 2024. PMID: 38935442
-
Unifying evolutionary and thermodynamic information for RNA folding of multiple alignments.Nucleic Acids Res. 2008 Nov;36(20):6355-62. doi: 10.1093/nar/gkn544. Epub 2008 Oct 4. Nucleic Acids Res. 2008. PMID: 18836192 Free PMC article.
-
RNAalifold: improved consensus structure prediction for RNA alignments.BMC Bioinformatics. 2008 Nov 11;9:474. doi: 10.1186/1471-2105-9-474. BMC Bioinformatics. 2008. PMID: 19014431 Free PMC article.
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
-
From consensus structure prediction to RNA gene finding.Brief Funct Genomic Proteomic. 2009 Nov;8(6):461-71. doi: 10.1093/bfgp/elp043. Brief Funct Genomic Proteomic. 2009. PMID: 19833701 Review.
Cited by
-
A Novel Small RNA, DsrO, in Deinococcus radiodurans Promotes Methionine Sulfoxide Reductase (msrA) Expression for Oxidative Stress Adaptation.Appl Environ Microbiol. 2022 Jun 14;88(11):e0003822. doi: 10.1128/aem.00038-22. Epub 2022 May 16. Appl Environ Microbiol. 2022. PMID: 35575549 Free PMC article.
-
Activation of an endogenous retrovirus-associated long non-coding RNA in human adenocarcinoma.Genome Med. 2015 Mar 5;7(1):22. doi: 10.1186/s13073-015-0142-6. eCollection 2015. Genome Med. 2015. PMID: 25821520 Free PMC article.
-
Computational analysis of RNA structures with chemical probing data.Methods. 2015 Jun;79-80:60-6. doi: 10.1016/j.ymeth.2015.02.003. Epub 2015 Feb 14. Methods. 2015. PMID: 25687190 Free PMC article. Review.
-
APeg3: regulation of Peg3 through an evolutionarily conserved ncRNA.Gene. 2014 May 1;540(2):251-7. doi: 10.1016/j.gene.2014.02.056. Epub 2014 Feb 28. Gene. 2014. PMID: 24582979 Free PMC article.
-
ProbeAlign: incorporating high-throughput sequencing-based structure probing information into ncRNA homology search.BMC Bioinformatics. 2014;15 Suppl 9(Suppl 9):S15. doi: 10.1186/1471-2105-15-S9-S15. Epub 2014 Sep 10. BMC Bioinformatics. 2014. PMID: 25253206 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

