Pax6 exerts regional control of cortical progenitor proliferation via direct repression of Cdk6 and hypophosphorylation of pRb
- PMID: 23622063
- PMCID: PMC3898967
- DOI: 10.1016/j.neuron.2013.02.012
Pax6 exerts regional control of cortical progenitor proliferation via direct repression of Cdk6 and hypophosphorylation of pRb
Abstract
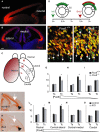
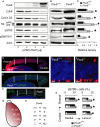
The mechanisms by which early spatiotemporal expression patterns of transcription factors such as Pax6 regulate cortical progenitors in a region-specific manner are poorly understood. Pax6 is expressed in a gradient across the developing cortex and is essential for normal corticogenesis. We found that constitutive or conditional loss of Pax6 increases cortical progenitor proliferation by amounts that vary regionally with normal Pax6 levels. We compared the gene expression profiles of equivalent Pax6-expressing progenitors isolated from Pax6⁺/⁺ and Pax6⁻/⁻ cortices and identified many negatively regulated cell-cycle genes, including Cyclins and Cdks. Biochemical assays indicated that Pax6 directly represses Cdk6 expression. Cyclin/Cdk repression inhibits retinoblastoma protein (pRb) phosphorylation, thereby limiting the transcription of genes that directly promote the mechanics of the cell cycle, and we found that Pax6 inhibits pRb phosphorylation and represses genes involved in DNA replication. Our results indicate that Pax6's modulation of cortical progenitor cell cycles is regional and direct.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Conditional activation of Pax6 in the developing cortex of transgenic mice causes progenitor apoptosis.Development. 2007 Apr;134(7):1311-22. doi: 10.1242/dev.02809. Epub 2007 Feb 28. Development. 2007. PMID: 17329367
-
Er81 is a downstream target of Pax6 in cortical progenitors.BMC Dev Biol. 2008 Feb 28;8:23. doi: 10.1186/1471-213X-8-23. BMC Dev Biol. 2008. PMID: 18307776 Free PMC article.
-
Controlled overexpression of Pax6 in vivo negatively autoregulates the Pax6 locus, causing cell-autonomous defects of late cortical progenitor proliferation with little effect on cortical arealization.Development. 2007 Feb;134(3):545-55. doi: 10.1242/dev.02764. Epub 2007 Jan 3. Development. 2007. PMID: 17202185 Free PMC article.
-
Transcriptional and epigenetic mechanisms of early cortical development: An examination of how Pax6 coordinates cortical development.J Comp Neurol. 2016 Feb 15;524(3):609-29. doi: 10.1002/cne.23866. Epub 2015 Aug 25. J Comp Neurol. 2016. PMID: 26304102 Free PMC article. Review.
-
Regulation of gene expression by Pax6 in ocular cells: a case of tissue-preferred expression of crystallins in lens.Int J Dev Biol. 2004;48(8-9):829-44. doi: 10.1387/ijdb.041866ac. Int J Dev Biol. 2004. PMID: 15558475 Free PMC article. Review.
Cited by
-
Silencing of lncRNA PKIA-AS1 Attenuates Spinal Nerve Ligation-Induced Neuropathic Pain Through Epigenetic Downregulation of CDK6 Expression.Front Cell Neurosci. 2019 Feb 20;13:50. doi: 10.3389/fncel.2019.00050. eCollection 2019. Front Cell Neurosci. 2019. PMID: 30873006 Free PMC article.
-
The role of the diencephalon in the guidance of thalamocortical axons in mice.Development. 2020 Jun 26;147(12):dev184523. doi: 10.1242/dev.184523. Development. 2020. PMID: 32541009 Free PMC article.
-
Molecular and cellular evolution of corticogenesis in amniotes.Cell Mol Life Sci. 2020 Apr;77(8):1435-1460. doi: 10.1007/s00018-019-03315-x. Epub 2019 Sep 28. Cell Mol Life Sci. 2020. PMID: 31563997 Free PMC article. Review.
-
Growth and folding of the mammalian cerebral cortex: from molecules to malformations.Nat Rev Neurosci. 2014 Apr;15(4):217-32. doi: 10.1038/nrn3707. Nat Rev Neurosci. 2014. PMID: 24646670 Free PMC article. Review.
-
Zic4-Lineage Cells Increase Their Contribution to Visual Thalamic Nuclei during Murine Embryogenesis If They Are Homozygous or Heterozygous for Loss of Pax6 Function.eNeuro. 2018 Oct 23;5(5):ENEURO.0367-18.2018. doi: 10.1523/ENEURO.0367-18.2018. eCollection 2018 Sep-Oct. eNeuro. 2018. PMID: 30406191 Free PMC article.
References
-
- Appolloni I., Calzolari F., Barilari M., Terrile M., Daga A., Malatesta P. Antagonistic modulation of gliomagenesis by Pax6 and Olig2 in PDGF-induced oligodendroglioma. Int. J. Cancer. 2012;131:E1078–E1087. - PubMed
-
- Asami M., Pilz G.A., Ninkovic J., Godinho L., Schroeder T., Huttner W.B., Götz M. The role of Pax6 in regulating the orientation and mode of cell division of progenitors in the mouse cerebral cortex. Development. 2011;138:5067–5078. - PubMed
-
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300.
-
- Biggin M.D. Animal transcription networks as highly connected, quantitative continua. Dev. Cell. 2011;21:611–626. - PubMed
-
- Bishop K.M., Goudreau G., O’Leary D.D. Regulation of area identity in the mammalian neocortex by Emx2 and Pax6. Science. 2000;288:344–349. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

