A Cdk7-Cdk4 T-loop phosphorylation cascade promotes G1 progression
- PMID: 23622515
- PMCID: PMC3677717
- DOI: 10.1016/j.molcel.2013.04.003
A Cdk7-Cdk4 T-loop phosphorylation cascade promotes G1 progression
Abstract
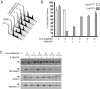
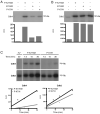
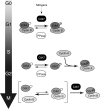
Eukaryotic cell division is controlled by cyclin-dependent kinases (CDKs), which require phosphorylation by a CDK-activating kinase (CAK) for full activity. Chemical genetics uncovered requirements for the metazoan CAK Cdk7 in determining cyclin specificity and activation order of Cdk2 and Cdk1 during S and G2 phases. It was unknown if Cdk7 also activates Cdk4 and Cdk6 to promote passage of the restriction (R) point, when continued cell-cycle progression becomes mitogen independent, or if CDK-activating phosphorylation regulates G1 progression. Here we show that Cdk7 is a Cdk4- and Cdk6-activating kinase in human cells, required to maintain activity, not just to establish the active state, as is the case for Cdk1 and Cdk2. Activating phosphorylation of Cdk7 rises concurrently with that of Cdk4 as cells exit quiescence and accelerates Cdk4 activation in vitro. Therefore, mitogen signaling drives a CDK-activation cascade during G1 progression, and CAK might be rate-limiting for R point passage.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures




References
-
- Aprelikova O, Xiong Y, Liu ET. Both p16 and p21 families of cyclin-dependent kinase (CDK) inhibitors block the phosphorylation of cyclin-dependent kinases by the CDK-activating kinase. J Biol Chem. 1995;270:18195–18197. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

