An archaeal RadA paralog influences presynaptic filament formation
- PMID: 23622866
- PMCID: PMC4084651
- DOI: 10.1016/j.dnarep.2013.03.003
An archaeal RadA paralog influences presynaptic filament formation
Abstract
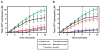
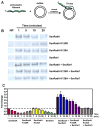
Recombinases of the RecA family play vital roles in homologous recombination, a high-fidelity mechanism to repair DNA double-stranded breaks. These proteins catalyze strand invasion and exchange after forming dynamic nucleoprotein filaments on ssDNA. Increasing evidence suggests that stabilization of these dynamic filaments is a highly conserved function across diverse species. Here, we analyze the presynaptic filament formation and DNA binding characteristics of the Sulfolobus solfataricus recombinase SsoRadA in conjunction with the SsoRadA paralog SsoRal1. In addition to constraining SsoRadA ssDNA-dependent ATPase activity, the paralog also enhances SsoRadA ssDNA binding, effectively influencing activities necessary for presynaptic filament formation. These activities result in enhanced SsoRadA-mediated strand invasion in the presence of SsoRal1 and suggest a filament stabilization function for the SsoRal1 protein.
Copyright © 2013 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



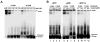


Similar articles
-
Characterization of an archaeal recombinase paralog that exhibits novel anti-recombinase activity.Mutat Res. 2020 May-Dec;821:111703. doi: 10.1016/j.mrfmmm.2020.111703. Epub 2020 May 7. Mutat Res. 2020. PMID: 32416400
-
A recombinase paralog from the hyperthermophilic crenarchaeon Sulfolobus solfataricus enhances SsoRadA ssDNA binding and strand displacement.Gene. 2013 Feb 15;515(1):128-39. doi: 10.1016/j.gene.2012.11.010. Epub 2012 Dec 6. Gene. 2013. PMID: 23220019 Free PMC article.
-
The single-stranded DNA binding protein of Sulfolobus solfataricus acts in the presynaptic step of homologous recombination.J Mol Biol. 2010 Mar 19;397(1):31-45. doi: 10.1016/j.jmb.2010.01.004. Epub 2010 Jan 18. J Mol Biol. 2010. PMID: 20080104
-
Divalent metal cofactors differentially modulate RadA-mediated strand invasion and exchange in Saccharolobus solfataricus.Biosci Rep. 2023 Feb 27;43(2):BSR20221807. doi: 10.1042/BSR20221807. Biosci Rep. 2023. PMID: 36601994 Free PMC article.
-
The RadA Recombinase and Paralogs of the Hyperthermophilic Archaeon Sulfolobus solfataricus.Methods Enzymol. 2018;600:255-284. doi: 10.1016/bs.mie.2017.12.009. Epub 2018 Feb 3. Methods Enzymol. 2018. PMID: 29458762
Cited by
-
Role of RadA and DNA Polymerases in Recombination-Associated DNA Synthesis in Hyperthermophilic Archaea.Biomolecules. 2020 Jul 14;10(7):1045. doi: 10.3390/biom10071045. Biomolecules. 2020. PMID: 32674430 Free PMC article.
-
Mediators of homologous DNA pairing.Cold Spring Harb Perspect Biol. 2014 Oct 9;6(12):a016451. doi: 10.1101/cshperspect.a016451. Cold Spring Harb Perspect Biol. 2014. PMID: 25301930 Free PMC article. Review.
-
Nanobiomotors of archaeal DNA repair machineries: current research status and application potential.Cell Biosci. 2014 Jun 25;4:32. doi: 10.1186/2045-3701-4-32. eCollection 2014. Cell Biosci. 2014. PMID: 24995126 Free PMC article. Review.
-
Commentary on: Divalent metal cofactors differentially modulate RadA-mediated strand invasion and exchange in Saccharolobus solfataricus.Biosci Rep. 2023 Jun 28;43(6):BSR20230058. doi: 10.1042/BSR20230058. Biosci Rep. 2023. PMID: 37334574 Free PMC article.
-
Flipping chromosomes in deep-sea archaea.PLoS Genet. 2017 Jun 19;13(6):e1006847. doi: 10.1371/journal.pgen.1006847. eCollection 2017 Jun. PLoS Genet. 2017. PMID: 28628615 Free PMC article.
References
-
- Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. - PubMed
-
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

