The budding yeast point centromere associates with two Cse4 molecules during mitosis
- PMID: 23623551
- PMCID: PMC3893307
- DOI: 10.1016/j.cub.2013.03.042
The budding yeast point centromere associates with two Cse4 molecules during mitosis
Abstract
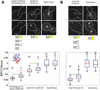
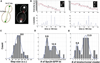
The centromere is defined by the incorporation of the centromere-specific histone H3 variant centromere protein A (CENP-A). Like histone H3, CENP-A can form CENP-A-H4 heterotetramers in vitro. However, the in vivo conformation of CENP-A chromatin has been proposed by different studies as hemisomes, canonical, or heterotypic nucleosomes. A clear understanding of the in vivo architecture of CENP-A chromatin is important, because it influences the molecular mechanisms of the assembly and maintenance of the centromere and its function in kinetochore nucleation. A key determinant of this architecture is the number of CENP-A molecules bound to the centromere. Accurate measurement of this number can limit possible centromere architectures. The genetically defined point centromere in the budding yeast Saccharomyces cerevisiae provides a unique opportunity to define this number accurately, as this 120-bp-long centromere can at the most form one nucleosome or hemisome. Using novel live-cell fluorescence microscopy assays, we demonstrate that the budding yeast centromere recruits two Cse4 (ScCENP-A) molecules. These molecules are deposited during S phase and they remain stably bound through late anaphase. Our studies suggest that the budding yeast centromere incorporates a Cse4-H4 tetramer.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures


References
-
- Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

