Evidence of microbial regulation of biogeochemical cycles from a study on methane flux and land use change
- PMID: 23624469
- PMCID: PMC3697577
- DOI: 10.1128/AEM.00095-13
Evidence of microbial regulation of biogeochemical cycles from a study on methane flux and land use change
Abstract
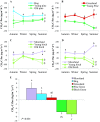
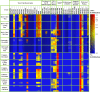
Microbes play an essential role in ecosystem functions, including carrying out biogeochemical cycles, but are currently considered a black box in predictive models and all global biodiversity debates. This is due to (i) perceived temporal and spatial variations in microbial communities and (ii) lack of ecological theory explaining how microbes regulate ecosystem functions. Providing evidence of the microbial regulation of biogeochemical cycles is key for predicting ecosystem functions, including greenhouse gas fluxes, under current and future climate scenarios. Using functional measures, stable-isotope probing, and molecular methods, we show that microbial (community diversity and function) response to land use change is stable over time. We investigated the change in net methane flux and associated microbial communities due to afforestation of bog, grassland, and moorland. Afforestation resulted in the stable and consistent enhancement in sink of atmospheric methane at all sites. This change in function was linked to a niche-specific separation of microbial communities (methanotrophs). The results suggest that ecological theories developed for macroecology may explain the microbial regulation of the methane cycle. Our findings provide support for the explicit consideration of microbial data in ecosystem/climate models to improve predictions of biogeochemical cycles.
Figures


References
-
- Gans J, Wolinsky M, Dunbar J. 2005. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387–1390 - PubMed
-
- Harris J. 2009. Soil microbial communities and restoration ecology: facilitators or followers? Science 325:573–574 - PubMed
-
- Isbell F, Calcagno V, Hector A, Connolly J, Harpole WS, Reich PB, Scherer-Lorenzen M, Schmid B, Tilman D, van Ruijven J, Weigelt A, Wilsey BJ, Zavaleta ES, Loreau M. 2011. High plant diversity is needed to maintain ecosystem services. Nature 477:199–202 - PubMed
-
- Reich PB, Tilman D, Isbell F, Mueller K, Hobbie SE, Flynn DF, Eisenhauer N. 2012. Impacts of biodiversity loss escalate through time as redundancy fades. Science 336:589–592 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

