Identification of transcriptional regulators in the mouse immune system
- PMID: 23624555
- PMCID: PMC3690947
- DOI: 10.1038/ni.2587
Identification of transcriptional regulators in the mouse immune system
Abstract
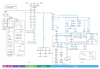
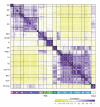
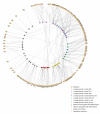
The differentiation of hematopoietic stem cells into cells of the immune system has been studied extensively in mammals, but the transcriptional circuitry that controls it is still only partially understood. Here, the Immunological Genome Project gene-expression profiles across mouse immune lineages allowed us to systematically analyze these circuits. To analyze this data set we developed Ontogenet, an algorithm for reconstructing lineage-specific regulation from gene-expression profiles across lineages. Using Ontogenet, we found differentiation stage-specific regulators of mouse hematopoiesis and identified many known hematopoietic regulators and 175 previously unknown candidate regulators, as well as their target genes and the cell types in which they act. Among the previously unknown regulators, we emphasize the role of ETV5 in the differentiation of γδ T cells. As the transcriptional programs of human and mouse cells are highly conserved, it is likely that many lessons learned from the mouse model apply to humans.
Figures




Comment in
-
Deconstructing development.Nat Immunol. 2013 Jun;14(6):529-31. doi: 10.1038/ni.2613. Nat Immunol. 2013. PMID: 23685815 No abstract available.
References
-
- Shay T, Jojic V, Zuk O, Rothamel K, Puyraimond-Zemmour D, Feng T, Wakamatsu E, Benoist C, Koller D, Regev A, ImmGen Consortium Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proceedings of the National Academy of Sciences. 2013;110(8):2946–2951. - PMC - PubMed
-
- Heng TSP, Painter MW, Elpek K, Lukacs-Kornek V, Mauermann N, Turley SJ, Koller D, Kim FS, Wagers AJ, Asinovski N, Davis S, Fassett M, Feuerer M, Gray DHD, Haxhinasto S, Hill JA, Hyatt G, Laplace C, Leatherbee K, Mathis D, Benoist C, Jianu R, Laidlaw DH, Best JA, Knell J, Goldrath AW, Jarjoura J, Sun JC, Zhu Y, Lanier LL, Ergun A, Li Z, Collins JJ, Shinton SA, Hardy RR, Friedline R, Sylvia K, Kang J. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9(10):1091–1094. - PubMed
-
- Iwasaki H, Akashi K. Myeloid Lineage Commitment from the Hematopoietic Stem Cell. Immunity. 2007;26(6):726–740. - PubMed
-
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, Frampton GM, Drake ACB, Leskov I, Nilsson B, Preffer F, Dombkowski D, Evans JW, Liefeld T, Smutko JS, Chen J, Friedman N, Young RA, Golub TR, Regev A, Ebert BL. Densely Interconnected Transcriptional Circuits Control Cell States in Human Hematopoiesis. Cell. 2011;144(2):296–309. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

