BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis
- PMID: 23624714
- PMCID: PMC3663277
- DOI: 10.1105/tpc.113.110684
BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis
Abstract
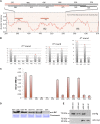
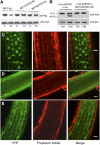
BINDING PROTEIN (BiP) is a major chaperone in the endoplasmic reticulum (ER) lumen, and this study shows that BiP binds to the C-terminal tail of the stress sensor/transducer bZIP28, a membrane-associated transcription factor, retaining it in the ER under unstressed conditions. In response to ER stress, BiP dissociates from bZIP28, allowing it to be mobilized from the ER to the Golgi where it is proteolytically processed and released to enter the nucleus. Under unstressed conditions, BiP binds to bZIP28 as it binds to other client proteins, through its substrate binding domain. BiP dissociates from bZIP28 even when bZIP28's exit from the ER or its release from the Golgi is blocked. Both BiP1 and BiP3 bind bZIP28, and overexpression of either BiP detains bZIP28 in the ER under stress conditions. A C-terminally truncated mutant of bZIP28 eliminating most of the lumenal domain does not bind BiP and is not retained in the ER under unstressed conditions. BiP binding sites in the C-terminal tail of bZIP28 were identified in a phage display system. BiP was found to bind to intrinsically disordered regions on bZIP28's lumen-facing tail. Thus, the dissociation of BiP from the C-terminal tail of bZIP28 is a major switch that activates one arm of the unfolded protein response signaling pathway in plants.
Figures







References
-
- Bechtold N., Ellis J., Pelletier G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316: 1194–1199
-
- Bertolotti A., Zhang Y., Hendershot L.M., Harding H.P., Ron D. (2000). Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2: 326–332 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

