On your histone mark, SET, methylate!
- PMID: 23625014
- PMCID: PMC3741215
- DOI: 10.4161/epi.24451
On your histone mark, SET, methylate!
Abstract
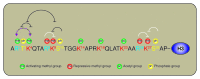
Lysine methylation of histones and non-histone proteins has emerged in recent years as a posttranslational modification with wide-ranging cellular implications beyond epigenetic regulation. The molecular interactions between lysine methyltransferases and their substrates appear to be regulated by posttranslational modifications surrounding the lysine methyl acceptor. Two very interesting examples of this cross-talk between methyl-lysine sites are found in the SET (Su(var)3-9, Enhancer-of-zeste, Trithorax) domain-containing lysine methyltransferases SET7 and SETDB1, whereby the histone H3 trimethylated on lysine 4 (H3K4 (me3) ) modification prevents methylation by SETDB1 on H3 lysine 9 (H3K9) and the histone H3 trimethylated on lysine 9 (H3K9 (me3) ) modification prevents methylation by SET7 on H3K4. A similar cross-talk between posttranslational modifications regulates the functions of non-histone proteins such as the tumor suppressor p53 and the DNA methyltransferase DNMT1. Herein, in cis effects of acetylation, phosphorylation, as well as arginine and lysine methylation on lysine methylation events will be discussed.
Keywords: chromatin; chromatin signaling; lysine methylation; lysine methyltransferase.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
