Functional properties and structural requirements of the plasmid pMV158-encoded MobM relaxase domain
- PMID: 23625844
- PMCID: PMC3697542
- DOI: 10.1128/JB.02264-12
Functional properties and structural requirements of the plasmid pMV158-encoded MobM relaxase domain
Abstract
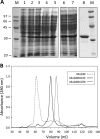
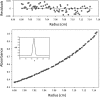
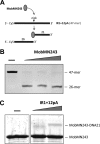
A crucial element in the horizontal transfer of mobilizable and conjugative plasmids is the relaxase, a single-stranded endonuclease that nicks the origin of transfer (oriT) of the plasmid DNA. The relaxase of the pMV158 mobilizable plasmid is MobM (494 residues). In solution, MobM forms a dimer through its C-terminal domain, which is proposed to anchor the protein to the cell membrane and to participate in type 4 secretion system (T4SS) protein-protein interactions. In order to gain a deeper insight into the structural MobM requirements for efficient DNA catalysis, we studied two endonuclease domain variants that include the first 199 or 243 amino acid residues (MobMN199 and MobMN243, respectively). Our results confirmed that the two proteins behaved as monomers in solution. Interestingly, MobMN243 relaxed supercoiled DNA and cleaved single-stranded oligonucleotides harboring oriTpMV158, whereas MobMN199 was active only on supercoiled DNA. Protein stability studies using gel electrophoresis and mass spectrometry showed increased susceptibility to degradation at the domain boundary between the N- and C-terminal domains, suggesting that the domains change their relative orientation upon DNA binding. Overall, these results demonstrate that MobMN243 is capable of nicking the DNA substrate independently of its topology and that the amino acids 200 to 243 modulate substrate specificity but not the nicking activity per se. These findings suggest that these amino acids are involved in positioning the DNA for the nuclease reaction rather than in the nicking mechanism itself.
Figures



Similar articles
-
Nicking activity of the pMV158 MobM relaxase on cognate and heterologous origins of transfer.Plasmid. 2013 Jul;70(1):120-30. doi: 10.1016/j.plasmid.2013.03.004. Epub 2013 Apr 3. Plasmid. 2013. PMID: 23562993
-
The MobM relaxase domain of plasmid pMV158: thermal stability and activity upon Mn2+ and specific DNA binding.Nucleic Acids Res. 2011 May;39(10):4315-29. doi: 10.1093/nar/gkr049. Epub 2011 Feb 3. Nucleic Acids Res. 2011. PMID: 21296755 Free PMC article.
-
The mobilization protein, MobM, of the streptococcal plasmid pMV158 specifically cleaves supercoiled DNA at the plasmid oriT.J Mol Biol. 1997 Mar 7;266(4):688-702. doi: 10.1006/jmbi.1996.0824. J Mol Biol. 1997. PMID: 9102462
-
The diversity of conjugative relaxases and its application in plasmid classification.FEMS Microbiol Rev. 2009 May;33(3):657-87. doi: 10.1111/j.1574-6976.2009.00168.x. FEMS Microbiol Rev. 2009. PMID: 19396961 Review.
-
Nicking by transesterification: the reaction catalysed by a relaxase.Mol Microbiol. 1997 Sep;25(6):1011-22. doi: 10.1046/j.1365-2958.1997.5241885.x. Mol Microbiol. 1997. PMID: 9350859 Review.
Cited by
-
Multiple plasmid origin-of-transfer regions might aid the spread of antimicrobial resistance to human pathogens.Microbiologyopen. 2020 Dec;9(12):e1129. doi: 10.1002/mbo3.1129. Epub 2020 Oct 27. Microbiologyopen. 2020. PMID: 33111499 Free PMC article.
-
Unraveling Genomic and Pathogenic Features of Aeromonas ichthyocola sp. nov., Aeromonas mytilicola sp. nov., and Aeromonas mytilicola subsp. aquatica subsp. nov.Animals (Basel). 2025 Mar 26;15(7):948. doi: 10.3390/ani15070948. Animals (Basel). 2025. PMID: 40218343 Free PMC article.
-
Bringing them together: plasmid pMV158 rolling circle replication and conjugation under an evolutionary perspective.Plasmid. 2014 Jul;74:15-31. doi: 10.1016/j.plasmid.2014.05.004. Epub 2014 Jun 2. Plasmid. 2014. PMID: 24942190 Free PMC article. Review.
-
Mobilizable Rolling-Circle Replicating Plasmids from Gram-Positive Bacteria: A Low-Cost Conjugative Transfer.Microbiol Spectr. 2014 Sep 19;2(5):8. doi: 10.1128/microbiolspec.PLAS-0008-2013. Microbiol Spectr. 2014. PMID: 25606350 Free PMC article.
-
Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.Proc Natl Acad Sci U S A. 2017 Aug 8;114(32):E6526-E6535. doi: 10.1073/pnas.1702971114. Epub 2017 Jul 24. Proc Natl Acad Sci U S A. 2017. PMID: 28739894 Free PMC article.
References
-
- de la Cruz F, Frost LS, Meyer RJ, Zechner EL. 2010. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 34:18–40 - PubMed
-
- Lanka E, Wilkins BM. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141–169 - PubMed
-
- Wozniak RAF, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8:552–563 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

