Helix unfolding/refolding characterizes the functional dynamics of Staphylococcus aureus Clp protease
- PMID: 23625918
- PMCID: PMC3682565
- DOI: 10.1074/jbc.M113.452714
Helix unfolding/refolding characterizes the functional dynamics of Staphylococcus aureus Clp protease
Abstract
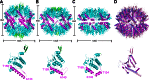
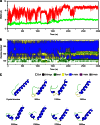
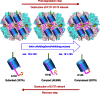
The ATP-dependent Clp protease (ClpP) plays an essential role not only in the control of protein quality but also in the regulation of bacterial pathogen virulence, making it an attractive target for antibacterial treatment. We have previously determined the crystal structures of Staphylococcus aureus ClpP (SaClpP) in two different states, extended and compressed. To investigate the dynamic switching of ClpP between these states, we performed a series of molecular dynamics simulations. During the structural transition, the long and straight helix E in the extended SaClpP monomer underwent an unfolding/refolding process, resulting in a kinked helix very similar to that in the compressed monomer. As a stable intermediate in the molecular dynamics simulation, the compact state was suggested and subsequently identified in x-ray crystallographic experiment. Our combined studies also determined that Ala(140) acted as a "hinge" during the transition between the extended and compressed states, and Glu(137) was essential for stabilizing the compressed state. Overall, this study provides molecular insights into the dynamics and mechanism of the functional conformation changes of SaClpP. Given the highly conserved sequences of ClpP proteins among different species, these findings potentially reflect a switching mechanism for the dynamic process shared in the whole ClpP family in general and thus aid in better understand the principles of Clp protease assembly and function.
Keywords: Bacterial Pathogenesis; Clp Protease; Computational Biology; Crystal Structure; Energy Landscape; Molecular Dynamics; Principle-Component Analysis; Protein Dynamics.
Figures



Similar articles
-
Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics.J Biol Chem. 2011 Oct 28;286(43):37590-601. doi: 10.1074/jbc.M111.277848. Epub 2011 Sep 7. J Biol Chem. 2011. PMID: 21900233 Free PMC article.
-
Reversible inhibition of the ClpP protease via an N-terminal conformational switch.Proc Natl Acad Sci U S A. 2018 Jul 10;115(28):E6447-E6456. doi: 10.1073/pnas.1805125115. Epub 2018 Jun 25. Proc Natl Acad Sci U S A. 2018. PMID: 29941580 Free PMC article.
-
Insights into structural network responsible for oligomerization and activity of bacterial virulence regulator caseinolytic protease P (ClpP) protein.J Biol Chem. 2012 Mar 16;287(12):9484-94. doi: 10.1074/jbc.M111.336222. Epub 2012 Jan 30. J Biol Chem. 2012. PMID: 22291011 Free PMC article.
-
Clp chaperones and proteases are central in stress survival, virulence and antibiotic resistance of Staphylococcus aureus.Int J Med Microbiol. 2014 Mar;304(2):142-9. doi: 10.1016/j.ijmm.2013.11.009. Epub 2013 Dec 1. Int J Med Microbiol. 2014. PMID: 24457183 Review.
-
Dynamics of the ClpP serine protease: a model for self-compartmentalized proteases.Crit Rev Biochem Mol Biol. 2014 Sep-Oct;49(5):400-12. doi: 10.3109/10409238.2014.925421. Epub 2014 Jun 10. Crit Rev Biochem Mol Biol. 2014. PMID: 24915503 Review.
Cited by
-
Functional Characterisation of ClpP Mutations Conferring Resistance to Acyldepsipeptide Antibiotics in Firmicutes.Chembiochem. 2020 Jul 16;21(14):1997-2012. doi: 10.1002/cbic.201900787. Epub 2020 Apr 9. Chembiochem. 2020. PMID: 32181548 Free PMC article.
-
Cryo-EM structure of the ClpXP protein degradation machinery.Nat Struct Mol Biol. 2019 Oct;26(10):946-954. doi: 10.1038/s41594-019-0304-0. Epub 2019 Oct 3. Nat Struct Mol Biol. 2019. PMID: 31582852 Free PMC article.
-
A gating mechanism for Pi release governs the mRNA unwinding by eIF4AI during translation initiation.Nucleic Acids Res. 2015 Dec 2;43(21):10157-67. doi: 10.1093/nar/gkv1033. Epub 2015 Oct 12. Nucleic Acids Res. 2015. PMID: 26464436 Free PMC article.
-
Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis.J Biol Chem. 2016 Apr 1;291(14):7465-76. doi: 10.1074/jbc.M115.700344. Epub 2016 Feb 8. J Biol Chem. 2016. PMID: 26858247 Free PMC article.
-
Structural Insights into Bortezomib-Induced Activation of the Caseinolytic Chaperone-Protease System in Mycobacterium tuberculosis.Nat Commun. 2025 Apr 11;16(1):3466. doi: 10.1038/s41467-025-58410-4. Nat Commun. 2025. PMID: 40216758 Free PMC article.
References
-
- Walsh C., Wright G. (2005) Introduction. Antibiotic resistance. Chem. Rev. 105, 391–394 - PubMed
-
- Williams K. J., Bax R. P. (2009) Curr. Opin. Biotechnol. 10, 157–163 - PubMed
-
- Sauer R. T., Bolon D. N., Burton B. M., Burton R. E., Flynn J. M., Grant R. A., Hersch G. L., Joshi S. A., Kenniston J. A., Levchenko I., Neher S. B., Oakes E. S., Siddiqui S. M., Wah D. A., Baker T. A. (2004) Sculpting the proteome with AAA+ proteases and disassembly machines. Cell 119, 9–18 - PMC - PubMed
-
- Wong P., Houry W. A. (2004) Chaperone networks in bacteria. Analysis of protein homeostasis in minimal cells. J. Struct. Biol. 146, 79–89 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

